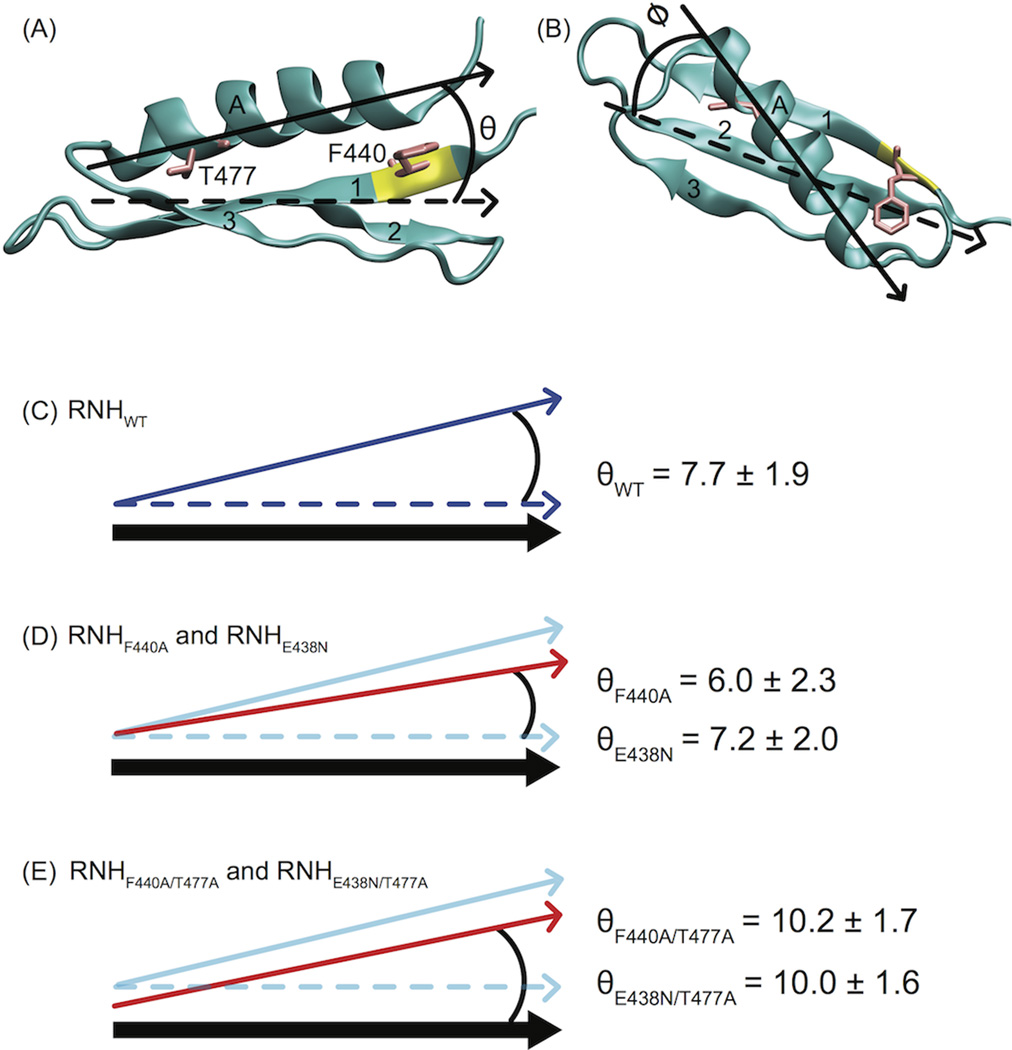
Figure 6.
Structural geometry for investigating crosstalk between sites 440 and 477 via MD. The orientation of α-helix A (Cα atoms) relative to the inertial axes of the beta sheet (β strands 1–3, backbone atoms listed in Table 4 is characterized by angles (A) θ and (B) φ. The average θ angle resulting from simulation of (C) RNHWT was 7.7 ± 1.9°. (D) The angle θ decreased slightly in the RNHF440A and RNHE438N mutants (red arrow) under MD simulation. (E) By contrast, RNHF440A/T477A and RNHE438N/T477A resulted in increased θ values. In (C) – (E), schematically, the thick black arrow represents the position of the β sheet. The blue solid and dashed arrows represent the position of α-helix A and the position of the β sheet inertial axis, respectively, for the RNHWT simulation. In (D) and (E), the red arrows indicate the position of α-helix A for the indicated simulations. In (D), because the simulation was not converged, the change of the angle is likely a transient effect of the conformational change within the 200 ns simulation. In (E), because the size of residue 477 decreases, the starting position of the helix is drawn in the cartoon differently from that in (D).