Figure 1.
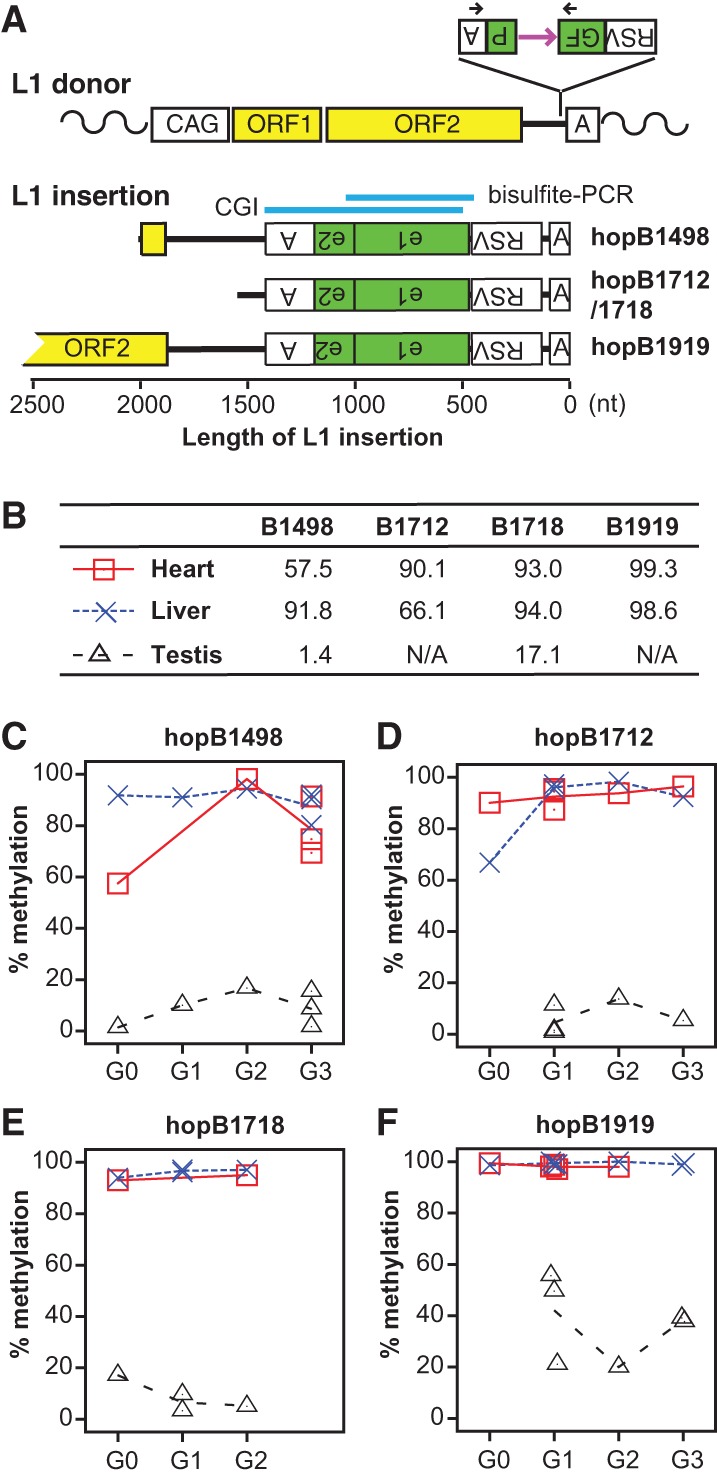
Integrating marker sequences into the mouse germline through L1 retrotransposition. (A) The donor L1 transgene ORFeus consists of a modified chicken beta-actin promoter (CAG), codon-optimized mouse L1 ORF1 and ORF2, a GFP-based retrotransposition indicator cassette in the 3′ UTR, and a polyadenylation signal (boxed letter A). The GFP cassette is placed in the antisense orientation relative to L1 transcription. The GFP reporter gene is flanked by Rous sarcoma virus (RSV) promoter and a polyadenylation signal but interrupted by a sense-oriented intron (purple horizontal arrow). Black arrows designate the location of genotyping primers. Retrotransposition creates a new L1 insertion, which is typically 5′ truncated, intronless, and trailed by a poly(A) DNA tract. Sequence structure of characterized L1 insertions is drawn to scale. All insertions are aligned at the 3′ end. For clarity, target site duplications and 3′ poly(A) tracts are omitted. Inverted letters indicate antisense orientation. For hopB1919, a near full-length insertion, only its 3′ portion is shown. Indicated above the insertions is the location of a 899-bp CGI predicted by newcpgseek as well as the region amplified by bisulfite PCR. (B) Methylation in GFP for all G0 animals. Animal IDs are indicated at the top. B1712 and B1718 are siblings with the same insertion. No testicular data are shown for B1919 and B1712 as both are female. (C–F) Contrasting methylation profiles between somatic tissues (heart and liver), and germ cells are maintained across multiple generations. Each point represents data from one individual animal. The connecting lines depict average methylation levels.
