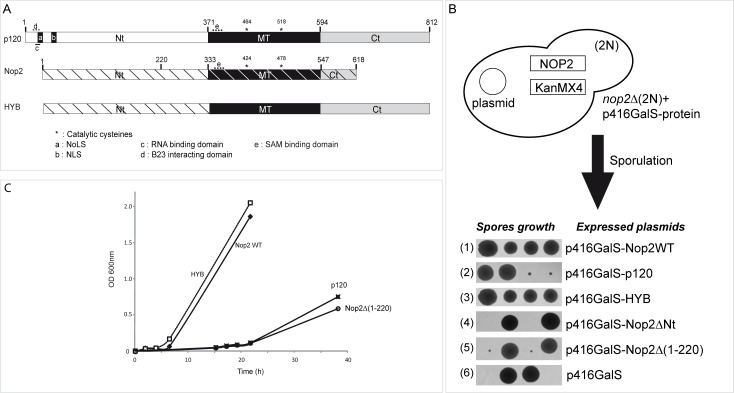
Fig 1. Protein domain structures of human p120 and S. cerevisiae Nop2 and conservation of the putative RNA:m5C-MTase active site.
(A) Global alignment of human p120, yeast Nop2 and HYB protein. Proteins are represented to scale and domains present in p120 and Nop2 as well as their positions are indicated (NTD: N-terminal domain, MTD: RNA:MTase catalytic domain, CTD: C-terminal domain). The positions of catalytic cysteine residues in the MTD are indicated by asterisks. Specific sequence motifs, identified in p120 and Nop2, are indicated by letters: a: p120 Nucleolar Localization Signal (NoLS), b: p120 Nuclear Localization Signal (NLS), c: RNA-binding domain, d: B23-interacting domain, e: AdoMet (SAM) binding domain. In HYB protein, p120 structural domains are indicated as grey and black boxes and the corresponding Nop2 domains are indicated as hatched white rectangle. (B) Preparation of haploid Nop2Δ strains by sporulation and tetrad dissection. Growth of individual spores expressing different variants is shown at the bottom. (C) Growth of viable complemented haploid strains in liquid YPG medium. Strains were inoculated at 0.05−0.1 units OD600 and grown for a maximum of 40 hours on a shaker at 30°C.