Abstract
Paenibacillus larvae is the causative agent of American foulbrood (AFB), the most serious honey bee brood bacterial disease. We isolated and characterized P. larvae-directed bacteriophages and developed criteria for safe phage therapy. Whole-genome analysis of a highly lytic virus of the family Siphoviridae (HB10c2) provided a detailed safety profile and uncovered its lysogenic nature and a putative beta-lactamase-like protein. To rate its antagonistic activity against the pathogens targeted and to specify potentially harmful effects on the bee population and the environment, P. larvae genotypes ERIC I to IV, representatives of the bee gut microbiota, and a broad panel of members of the order Bacillales were analyzed for phage HB10c2-induced lysis. Breeding assays with infected bee larvae revealed that the in vitro phage activity observed was not predictive of the real-life scenario and therapeutic efficacy. On the basis of the disclosed P. larvae-bacteriophage coevolution, we discuss the future prospects of AFB phage therapy.
INTRODUCTION
The widespread collapse of honeybee (Apis mellifera) colonies has led to serious concerns worldwide. Although the exact causes are not fully understood, infectious agents obviously pose a significant threat to bee health and may contribute to the decline of this major pollinator of crops, fruit, and wild plants. Despite increased bee mortality rates, adequate or authorized medical products to treat bacterial and viral bee infections are missing. This also holds true for American foulbrood (AFB), which is the most contagious and destructive bacterial infection affecting honey bees. The etiological agent is Paenibacillus larvae, a Gram-positive, rod-shaped, spore-forming bacterium. The extremely resilient and long-lived endospores are the infectious form, and only bee larvae at an age of less than 30 h are susceptible (1). Oral uptake of about 10 spores is sufficient to initiate a fatal infection in bee larvae. After germination, P. larvae proliferates in the midgut, breaches the epithelium, and invades the hemocoel of bee larvae. This invasion coincides with the death of infected larvae, which turn into a brown, glue-like liquid that contains vast amounts of P. larvae spores. Killed larvae are detected by nurse bees, which clean the cells and prepare them for new brood. During this cleaning, they incorporate spores of P. larvae and feed those to healthy larvae that subsequently become infected (2).
Different methods of treating AFB-infected colonies are known, including burning down the colonies, artificial swarming, and decontamination of the hives with NaOH (2). In some countries, the antibiotics oxytetracycline and tylosin are used prophylactically or to treat symptoms (3, 4). However, all known strategies exhibit severe disadvantages for beekeepers, bee colonies, and the environment. The registered use of antibiotics, e.g., has been withdrawn in many countries since antibiotic residues appear within the honey. European Community (EC) legislation (EC regulation 2377/90) limits the presence of antibiotics in honey, excluding their use for AFB therapy. Moreover, the accumulation of antibiotic resistance in the gut microbiota of bees has been detected. As proposed recently, this resistance might additionally lead to bee colony collapse (4).
Taken together, the evidence shows that new strategies for the control of AFB are urgently needed but difficult to develop. The introduction of new antibiotics in the multidrug resistance era may only reproduce known drawbacks. The application of antagonistic bacteria combating P. larvae seems not to be in sight. Also, essential oils have not proven to be effective enough in practical use (2). In addition, they may have disadvantageous side effects on bees or honey. Since bacteriophages, viruses that infect and lyse bacteria, have already shown great efficacy in controlling bacterial infections in humans and animals, phage therapy seems to be a compelling alternative for the following reasons (5, 6). First, phages as bactericidal agents can easily be discovered and have been used to treat human infections since the early 1900s. Second, phages are autodosing at the site of infection and show negligible inherent toxicity (7). Third, because of their host specificity, phages usually exhibit minimal disruption of the normal flora. Fourth, as phages eliminate bacteria by mechanisms that differ from those of antibiotics, cross-resistance is not observed (8). Fifth, since phages are isolated from the environment, they can be regarded as natural products with only an irrelevant environmental impact compared to antibiotic drugs causing sustainable resistance loads (7).
Nonetheless, the safety profile of therapeutic phages needs to be addressed. It is important to ensure that the phages selected do not exhibit generalized transduction or possess gene sequences with significant homology to antibiotic resistance genes and genes for other bacterial virulence factors (9, 10). Moreover, it is critical that the therapeutic phages lyse relevant strains of the targeted bacteria and spare bacteria of the bee microbiome and the habitat of the hives.
In this study, we isolated several P. larvae-specific bacteriophages and rigorously characterized the novel phage HB10c2 with respect to morphology, genomic potential, and host range. Moreover, we analyzed the potential of HB10c2 as a therapeutic agent in cocultivation assays with P. larvae and in in vitro breeding assays in which infected bee larvae were treated with this bacteriophage. Finally, we discuss prerequisites for successful and safe phage therapy of AFB in honey bees.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
All of the bacterial strains used in this study are listed in Table 1. The growth media and cultivation conditions used were as described elsewhere (https://www.dsmz.de/). P. larvae strains were cultivated on Columbia sheep blood agar (BD) at 37°C with 5% CO2. For liquid culture, P. larvae strains were grown in brain heart infusion (BHI) medium (37 g of BHI [Roth], 3 g of yeast extract [BD], 1 liter of H2O) at 37°C and 200 rpm. Field isolates of P. larvae ERIC I and II were derived from honey samples by the Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit (LAVES) Institute for Bee Research, Celle, Germany. Plaque assays were performed with the small-drop plaque assay system (12).
TABLE 1.
Lytic activity of bacteriophage HB10c2 against P. larvae, microbiota, and environmental Bacillales
| Strain | Gram staining | Lytic activity of phage HB10c2 |
|---|---|---|
| Paenibacillus larvae ERIC I | ||
| DSM 7030 | + | + |
| Isolate 5 | + | + |
| Isolate 9 | + | + |
| Isolate 11 | + | + |
| Isolate 15 | + | + |
| Isolate 22 | + | + |
| Isolate 24 | + | + |
| Isolate 25 | + | + |
| Isolate 26 | + | + |
| Isolate 29 | + | + |
| Isolate 145 | + | + |
| Isolate 146 | + | + |
| Isolate174 | + | + |
| Isolate 148 | + | + |
| Isolate 153 | + | + |
| Isolate 155 | + | + |
| Isolate 157 | + | + |
| Isolate 159 | + | + |
| Isolate 160 | + | + |
| Isolate 162 | + | + |
| Paenibacillus larvae ERIC II | ||
| DSM 16116 | + | + |
| DSM 25430 | + | + |
| Isolate 1 | + | + |
| Isolate 3 | + | + |
| Isolate 6 | + | + |
| Isolate 7 | + | + |
| Isolate 17 | + | + |
| Isolate 20 | + | + |
| Isolate 23 | + | + |
| Isolate 27 | + | + |
| Isolate 28 | + | + |
| Isolate 135 | + | + |
| Isolate 137 | + | + |
| Isolate 144 | + | + |
| Isolate 149 | + | + |
| Isolate 152 | + | + |
| Isolate 156 | + | + |
| Isolate 158 | + | + |
| Isolate 161 | + | + |
| Isolate 168 | + | + |
| Bacillus licheniformisa LMG 7559 | + | − |
| Bacillus megateriuma LMG 7127 | + | − |
| Bacillus pumilusa LMG 3455 | + | − |
| Bacillus subtilisa LMG 2099 | + | − |
| Bacillus borstelensisa DSM 6347 | + | − |
| Delftia acidovoransa LMG 1226 | − | − |
| Enterococcus faecalisa DSM 20376 | + | − |
| Janthinobacterium lividuma LMG 2892 | − | − |
| Kocuria roseaa DSM 20447 | + | − |
| Pseudomonas fluorescensa DSM 6147 | − | − |
| Staphylococcus pasteuria DSM 30868 | + | − |
| Streptococcus salivariusa DSM 20067 | + | − |
| Streptomyces griseusa DSM 1471 | + | − |
| Comamonas denitrificansa,b LMG 21602 | − | − |
| Salmonella entericaa,b DSM 11320 | − | − |
| Gluconobacter oxydansb DSM 2003 | − | − |
| Lactobacillus crispatusb LMG 9479 | + | − |
| Pedobacter africanusb LMG 10345 | − | − |
| Planomicrobium okeanokoitesb DSM 15489 | + | − |
| Ralstonia pickettiib LMG 5942 | − | − |
| Saccharibacter floricolab LMG 23170 | − | − |
| Paenibacillus amylolyticusc DSM 11730 | + | − |
| Brevibacillus brevisc DSM 30 | + | + |
| Brevibacillus laterosporusc DSM 25 | + | − |
| Paenibacillus pabulic DSM 3036 | + | − |
| Paenibacillus peoriaec DSM 8320 | + | − |
| Paenibacillus phyllosphaeraec DSM 17399 | + | + |
| Paenibacillus polymyxac DSM 36 | + | − |
| Paenibacillus popilliaec DSM 22700 | + | − |
| Paenibacillus xylanilyticusc DSM 17225 | + | − |
| Paenibacillus solic DSM 21316 | + | + |
| Paenibacillus humicusc DSM 212785 | + | + |
| Paenibacillus apiariusc DSM 5581 | + | − |
| Paenibacillus cookiic DSM 16944 | + | + |
| Paenibacillus elgiic DSM 22254 | + | − |
| Paenibacillus filicisc DSM 23916 | + | + |
| Paenibacillus fonticolac DSM 21315 | + | − |
| Paenibacillus lactisc DSM 15596 | + | + |
| Paenibacillus lautusc DSM 3035 | + | − |
| Paenibacillus nanensisc DSM 22867 | + | − |
| Paenibacillus alveic DSM 29 | + | + |
| Paenibacillus castaneaec DSM 19417 | + | + |
Microbiota of bee larvae.
Microbiota of adult bees.
Environmental Bacillales.
Isolation of phage HB10c2.
Bacteriophage HB10c2 was isolated from the glue-like liquid of a beehive with clinical symptoms caused by P. larvae ERIC I. The combs of the hives were stored at the LAVES Institute for Bee Research in Celle, Germany, at 4°C. The glue-like liquid was taken out of the comb, transferred into 50 ml of H2O, and dissolved for 48 h at 4°C. To kill contaminants, 5% (vol/vol) CHCl3 (VWR) was added; this was followed by centrifugation at 4,000 × g for 15 min. The supernatant was mixed with the same volume of 2× BHI medium and inoculated with P. larvae DSM 16116, representing ERIC II. The culture was grown for 3 days at 37°C and 200 rpm. After incubation with 5% (vol/vol) CHCl3, bacteria were sedimented by centrifugation at 4,000 × g for 15 min. Phages were purified from the supernatant by ultracentrifugation at 100,000 × g for 1 h at 4°C. Bacteriophages were dissolved in 500 μl of SM buffer (100 mM NaCl [Roth], 8 mM MgSO4·7H2O [Roth], 50 mM Tris [pH 7,5] [Roth], 0.002% [wt/vol] gelatin [Roth]).
Enrichment of phages.
P. larvae was grown on Columbia blood agar (Oxoid) at 37°C and 5% CO2 for 16 h. Bacteria were suspended in sterile H2O and centrifuged for 5 min at 5,000 × g. Sedimented bacteria were resuspended in double-distilled H2O to an optical density at 600 nm (OD600) of 15. For bacteriophage enrichment, 1 volume of the suspension was added to 0.5 volume of phage lysate and transferred into 1 volume of 2× BHI. After adsorption for 15 min at room temperature, the complete sample was added to 400 volumes of BHI and incubated overnight at 37°C and 200 rpm. To lyse the bacteria, 15 volumes of CHCl3 was added. The mixture was inverted and centrifuged at 3,000 × g for 5 min. Supernatant was transferred into sterile tubes with a drop of CHCl3 to inhibit bacterial growth. The lysate derived was used for further experiments.
Determination of viral burst size.
A volume of 400 ml of BHI was inoculated with 5 × 108 cells of P. larvae (DSM 25430) and 1.25 × 107 PFU of phage HB10c2. The culture was grown at 37°C and agitated at 200 rpm. A culture without phage HB10c2 was used as a control. After incubation, aliquots of each culture were taken and the number of bacterial cells was determined. For phage isolation, the cultures were mixed with 5% (vol/vol) CHCl3 and shaken for 15 min at 37°C. The aqueous phase was centrifuged at 12,000 × g for 15 min, and the supernatant was used for enumeration of bacteriophages in titration assays with the small-drop plaque assay system (12). The specific burst size of bacteriophage HB10c2 was calculated with the equation BHB10c2 = log(PFU0/PFU)/log(GP. larvae), where the burst size (B) was calculated by the PFU at time zero (PFU0) and the PFU after incubation (PFU) with regard to the number of host generations (G).
Electron microscopy analysis.
Phage HB10c2 was enriched in a total volume of 400 ml of BHI. Bacterial DNA was hydrolyzed by incubation with 10 U of DNase I (Macherey-Nagel) and 40 μg of RNase A (Sigma) for 30 min at room temperature. After the addition of NaCl (Roth) to a final concentration of 0.5 M and incubation on ice for 15 min, the phages were separated from bacterial debris by centrifugation at 8,000 × g for 20 min at 4°C. Phages were enriched by polyethylene glycol (PEG) 3000 (Roth) purification (13). The enriched phages were concentrated by CsCl equilibrium centrifugation at 100,000 × g for 12 h at 4°C after the addition of 10 g of CsCl (Roth) to the PEG-purified phages. Phages were allowed to adsorb to carbon film, rinsed with TE buffer (10 mM Tris-HCl, pH 8.0 [Roth], 2 mM EDTA [Sigma]), rinsed in double-distilled H2O, and negatively stained with 2% (wt/vol) aqueous uranyl acetate (Merck). Carbon films were then collected from the uranyl acetate drops with copper grids (300 mesh) and blotted dry with filter paper. After being air dried, samples were examined in a Zeiss TEM 910 at an acceleration voltage of 80 kV. Images were recorded digitally with a Slow-Scan charge-coupled device camera (ProScan; 1,024 by 1,024 pixels) and ITEM software (Olympus Soft Imaging Solutions). Image contrast and brightness were adjusted with Adobe Photoshop.
Phage sequencing.
Phages derived by CsCl equilibrium centrifugation were used for isolation of DNA via phenol-chloroform extraction, followed by chloroform extraction. DNA was precipitated with ethanol by adding 300 μl of 3 M sodium acetate (Roth). SMRTbell template libraries were prepared according to instructions from Pacific Biosciences, Menlo Park, CA, USA, following the procedure and checklist for low-input 10-kb template preparation and sequencing by using C2-C2 chemistry as described previously (14).
Genome analyses.
Phage genome assemblies were prepared by using the RS_HGAP_Assembly.3 protocol included in SMRTPortal version 2.2.0 and using standard parameters. For all phages, one final contig could be obtained, which was linearized because of recognition of distinct start and end points in the phage assemblies. A quality check of the final phage genomes regarding overall coverage, as well as single-nucleotide polymorphisms, was performed with SMRT View and Integrative Genomics Viewer (16). All phage genomes were annotated with Prokka 1.8 (PMID 24642063) with subsequent manual curation in Artemis (15).
Phage host range determination.
Bacterial susceptibility to phage HB10c2 was assayed with the small-drop plaque assay system (see above) and OD measurements in liquid culture. In brief, precultures of P. larvae were grown overnight in BHI medium. For host range determination, 2.4 × 107 bacterial cells in 50 ml of BHI were inoculated with 4 × 109 PFU of phage HB10c2. Infected cultures were incubated at 37°C and agitated at 200 rpm for 24 h. OD600 was determined every hour. Growth curves were depicted by nonlinear regression (18) by using the model f(t) = (N0 × K × er × t)/(K + N0 × er × t − 1), where N0 is the OD600 at time (t) zero, K is the maximum OD600 (after 24 h), and r is the expansion rate.
Field isolates of P. larvae ERIC I and II were grown on Columbia sheep blood agar, and phage susceptibility was tested in plaque assays. The identities of all field isolates to the species level and their ERIC genotypes were determined by 16S rRNA PCR and repetitive element sequence-based PCR (rep-PCR) as described elsewhere (17).
The gut microbiotas of honey bee larvae and adult bees were grown in their respective standard culture media as described at https://www.dsmz.de/. After resuspension in H2O, 1 × 106 bacterial cells were used for inoculation of 4 ml of BHI top agar in plaque assays. Plaque formation by phage HB10c2 was monitored for 2 days.
Phage therapy of A. mellifera.
The in vivo therapeutic effect of phage HB10c2 on P. larvae ERIC I- and II-infected bee larvae was analyzed in breeding assays as described previously (19, 40). Larvae <24 h old were transferred to 20 μl of diet A (50% royal jelly and 50% aqueous solution containing 2% yeast extract, 12% glucose, and 12% fructose) (day 1). On day 3, the larvae were fed 20 μl of diet B (50% royal jelly and 50% aqueous solution containing 3% yeast extract, 15% glucose, and 15% fructose). Increasing amounts (30, 40, and 50 μl) of diet C (50% royal jelly and 50% aqueous solution containing 4% yeast extract, 18% glucose, and 18% fructose) were fed from day 4 to day 6, respectively. Three replicates of 48 larvae from different, nonrelated queens were used for infection. Each larva was infected with 20 μl of diet A containing P. larvae spores (P. larvae ERIC I DSM 7030 or ERIC II DSM 25430 at 500 CFU/larva). To confirm P. larvae as the causative agent of the bee deaths observed, dead bee larvae were homogenized and plated on blood agar. The identity of P. larvae colonies was confirmed by 16S rRNA PCR (20). To ensure that phage HB10c2 has no effect on the mortality rate of developing A. mellifera, 50,000 PFU of phage HB10c2 were fed diet A. For phage therapy, bees were fed diet A containing P. larvae spores (500 CFU/larva) and bacteriophages (50,000 PFU/larva). Numbers of insects killed were used for statistical analysis. The significance of changes in the mortality rates was calculated by unpaired Student t test.
Nucleotide sequence accession number.
The genome sequence of bacteriophage HB10c2 was deposited in the GenBank database under accession number KP202972.
RESULTS
Phage isolation, propagation, and host cell lysis.
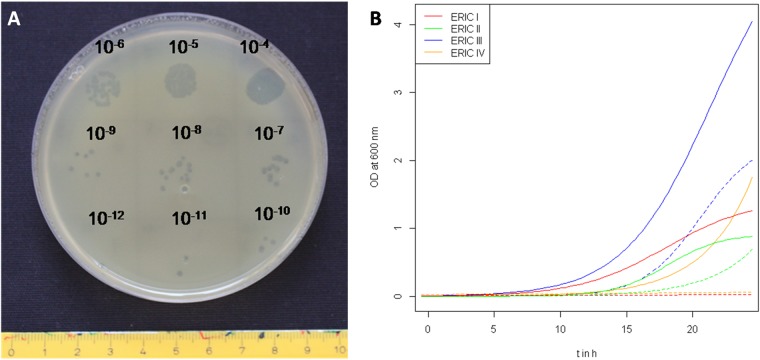
P. larvae-specific phages were isolated from infected beehives by plaque formation on lawns of P. larvae DSM 16116 (genotype ERIC II). Phage HB10c2, the most active isolate with regard to plaque size and clarity, originated from the glue-like larval remains of a beehive with clinical symptoms of AFB caused by P. larvae genotype ERIC I. The lytic activity of phage HB10c2 was further characterized after enrichment and subcultivation of phage lysates by titration assays (Fig. 1A). The calculated burst size of bacteriophage HB10c2 (BHB10c2) after infection of P. larvae ERIC II (DSM 25430) was 8.34. According to previous studies (21, 22) the particle-to-PFU ratio is about circa 15 bacteriophages per PFU, which suggests the release of approximately 125 phage particles per infected host cell. In addition to plaque assays and burst size calculation, the influence of phage HB10c2 on the growth of the four P. larvae genotypes, ERIC I to IV, was determined in liquid culture medium, respectively (Fig. 1B). The growth of P. larvae ERIC I and IV in coculture with HB10c2 was completely inhibited, whereas ERIC II and III exhibited a 50% lower cell density (OD600) than the control without bacteriophages.
FIG 1.
Plaque formation and host cell lysis by P. larvae-specific bacteriophage HB10c2. (A) Titration assay to determine the lytic activity of phage HB10c2 against P. larvae genotype ERIC II on top agar. (B) Growth of P. larvae ERIC I to IV (continuous lines) and effect of phage HB10c2 on bacterial growth (dotted lines) in BHI liquid medium. Phage HB10c2 had a significant effect (unpaired Student t test, P < 0.05) on the growth of ERIC I (P = 1.91 × 10−10), ERIC II (P = 3.06 × 10−7), ERIC III (P = 1.72 × 10−8), and ERIC IV (P = 7.12 × 10−4).
Electron microscopy reveals the B2 morphotype of phage HB10c2.
For a first classification of P. larvae-specific phage HB10c2, transmission electron microscopy of negatively stained phages was used. After phage purification by CsCl equilibrium centrifugation of P. larvae lysates, the morphology of the virions could be determined. Figure 2 shows representative intact phages with a capsid (length, 98.54 ± 0.54 nm; diameter, 46.79 ± 0.56 nm), a siphon (length, 165.34 ± 19.09 nm; diameter, 10.32 ± 0.21 nm), and a base plate (diameter, 22.77 ± 0.62 nm). According to these morphological properties, especially the length-to-width ratio, bacteriophage HB10c2 exhibited the B2 morphotype of the Siphoviridae family (23, 24).
FIG 2.
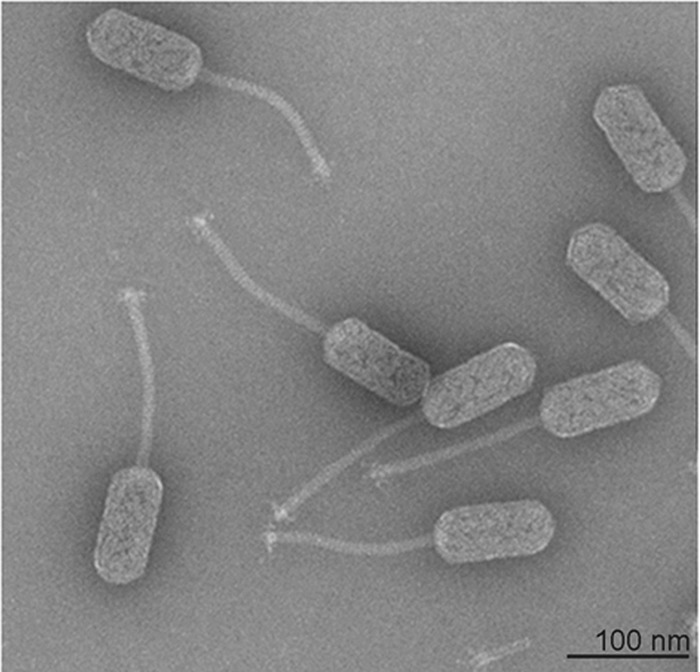
Transmission electron micrograph of bacteriophage HB10c2. Morphology of phage HB10c2 after negative staining and transmission electron microscopy. According to the average capsid, siphon, and base plate sizes, phage HB10c2 exhibits the B2 morphotype of the Siphoviridae family.
Whole-genome analysis of P. larvae phage HB10c2.
For further characterization with respect to gene content and genomic organization, the complete genome of phage HB10c2 was sequenced with the PacBio RS II system. The analysis revealed that the genome contains linear, double-stranded DNA with a length of 35,644 bp and a G+C content of 41.8%. We performed automatic annotation with Prokka (25) for the positioning of coding sequences (CDS) on three different start codons (ATG, GTG, and TTG) and the identification of putative Shine-Dalgarno sequences upstream of the coding regions. Accordingly, we determined 56 CDS for phage HB10c2 (52 ATG, 2 GTG, and 2 TTG), resulting in a coding percentage of 91.3%. Automatic annotation was manually refined with Artemis (15) and BLASTP. The genome could be divided into different functional clusters with genes, e.g., for structural proteins, replication, host lysis, or lysogeny (Fig. 3). Notably, we identified a gene encoding a putative beta-lactamase-like protein in phage HB10c2, which demonstrates the importance of genome sequencing for the risk assessment of putative therapeutic phages.
FIG 3.
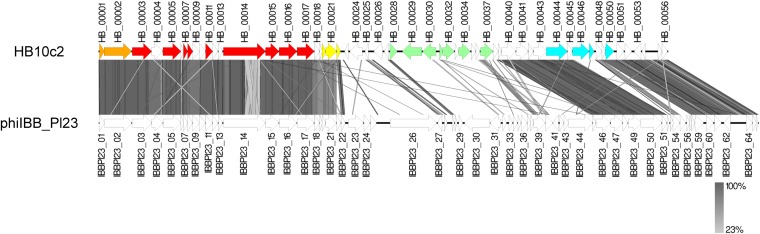
Comparison of the genomic structures of phages HB10c2 and phiBB_P123. The functional cluster for DNA packaging is orange, structural genes are red, replication genes are blue, host lysis genes are yellow, and genes involved in lysogeny are green. The image shown was generated by using Easyfig with amino acid sequence comparison. The range of amino acid identity levels is shown in the gradient scale.
At the right end of the phage chromosome, we identified a cluster of genes similar to Paenibacillus phage phiIBB_Pl23 (GenBank accession no. NC_021865) that show partial similarities to genes involved in replication. Among them were genes for a helicase (HB_00044), a nucleoside triphosphate pyrophosphohydrolase (HB_00047), and a RuvC-like protein (HB_00050). Apart from these genes, HB_00046 revealed similarities to a cytosine-C5-specific DNA methylase that might help to protect the phage DNA against degradation by the restriction system of the bacterial host. Aside from these genes, we could not identify a gene for a DNA polymerase, a primase, or a single-stranded binding protein that is essential for replication and may therefore be provided by the phage host.
As in most phage genomes, the genes for the small and large terminase (HB_00002) subunits of phage HB10c2 could be clearly identified upstream of the cluster of structural genes. In HB10c2, this cluster was identified at the left end of the chromosome. BLASTP analysis revealed the conserved domains Terminase_4 (pfam05119) and COG4626, respectively. Both proteins are identical to those from Paenibacillus phage phiIBB_Pl23. This cluster of DNA packaging genes is followed by the cluster of structural protein genes. All of these genes also show high similarities or are even identical to genes from Paenibacillus phage phiIBB_PI23. Among others, this cluster contains genes for a portal protein, a Clp protease-like protein (HB_00004), a tail length tape measure protein (HB_00014), and several structural proteins for the phage tail and head (HB_00014 to HB_00017).
Analysis with TMHMM Server v. 2.0 (26) revealed that the deduced amino acid sequences of HB_00020 and HB_00022 contain one and two transmembrane regions, respectively. Therefore, HB_00022 could be assigned to holin class II (27). Between the two putative holin genes, we identified a gene that is identical to a gene for an N-acetylmuramoyl l-alanine amidase from P. larvae ERIC I strain DSM 25719.
Downstream of the lysis cluster of phage HB10c2, we determined some genes located on the other strand that show no similarity to P. larvae phage phiIBB_Pl23 but do show similarity to genes of P. larvae ERIC I strain DSM 25719 (BioProject accession no. PRJNA42203) with unknown functions. Downstream of those genes, we identified CDS for an integrase (HB_00029), an excisionase-like protein (HB_000333), and antirepressor proteins (HB_00032 and HB_00037) and some genes that might play a role in the further regulation of the lysogenic state (HB_00030, HB_00031, and HB_00034). Taken together, the genome sequencing results suggest a potential for phage-mediated horizontal gene transfer.
Lytic activity of phage HB10c2 against different P. larvae genotypes and field isolates, members of the order Bacillales, and gut microbiota of A. mellifera.
To analyze the host range of phage HB10c2 and to exclude harmful effects on the bee gut microbiota or the environment, a broad panel of bacteria was tested for susceptibility to phage HB10c2-induced lysis by plaque assay. In addition to the four different known genotypes of P. larvae (ERIC I, DSM 7030; ERIC II, DSM 25430; ERIC III, LMG 16252; ERIC IV, LMG 16247) 20 ERIC I and II field isolates were tested. As shown in Table 1, all 40 isolates tested were sensitive to phage HB10c2-induced lysis.
In order to explore health implications for honey bees and possible ecological effects after assumptive therapy with phage HB10c2, representative culturable members of the larval and adult gut microbiotas of A. mellifera (28) and different members of the order Bacillales were analyzed in plaque assay (Table 1). In general, bacteriolytic activity of phage HB10c2 for Gram-negative bacteria was not observed. All of the species that allowed plaque formation belonged to the order Bacillales, and most of them were in the genus Paenibacillus. Interestingly, nine environmental members of the order Bacillales (Bacillus brevis, Paenibacillus phyllosphaerae, Paenibacillus soli, Paenibacillus humicus, Paenibacillus cookii, Paenibacillus filicis, Paenibacillus lactis, Paenibacillus alvei, and Paenibacillus castaneae) were susceptible to phage HB10c2-mediated lysis. This reveals that HB10c2 is a phage with a rather broad host range.
Treatment of P. larvae-infected bee larvae with phage HB10c2.
Since phage HB10c2 exhibited a strong lytic effect on P. larvae on agar plates and in liquid medium, we analyzed the in vivo therapeutic effect by using P. larvae ERIC I- and II-infected larvae of A. mellifera, respectively (Fig. 4). After feeding with a larval diet containing spores of P. larvae and phage HB10c2, the mortality rate of the developing bee larvae was analyzed daily for 14 days. In control experiments, bee larvae were fed only a larval diet or a larval diet containing phage HB10c2.
FIG 4.
Treatment of P. larvae-infected A. mellifera larvae with phage HB10c2. Bee larvae infected with P. larvae genotypes ERIC I (DSM 7030) and II (DSM 25430) exhibited a mortality rate of 100% after 14 days. Uninfected bee larvae (control) showed a mortality rate significantly lower than that of infected larvae (unpaired Student t test, P < 0.05). Uninfected larvae treated with phage HB10c2 had a mortality rate not significantly different from that of uninfected bee larvae without phage treatment (P = 0.2992). The mortality rate of P. larvae ERIC I- and II-infected bee larvae did not decline significantly after phage treatment (P = 0.4226 and P = 0.677, respectively). [d] p.i., days postinfection.
Larvae fed the larval diet alone exhibited a mortality rate of 25.00% ± 3.61%. The addition of phage HB10c2 to uninfected larvae led to a mortality rate of 31.25% ± 8.33%. Since the mortality rates of the two control groups are not significantly different (P = 0.2992), we concluded that phage HB10c2 has no harmful impact on the survival of bee larvae. In contrast, larvae infected with either P. larvae ERIC I or II had a mortality rate of 100% after 14 days. This significant difference from the respective control groups (P = 3.56 × 10−6) demonstrates the devastating effect of this pathogen. Larvae that were infected with P. larvae ERIC I or II and treated with phage HB10c2 showed mortality rates of 99.31% ± 1.20% and 98.61% ± 2.41%, respectively. Both values are not significantly different from the mortality rate of infected bee larvae without phage therapy (P = 0.4226 and P = 0.6770). Thus, we conclude that the application of phage HB10c2 did not exhibit therapeutic effects on bee larvae suffering from AFB.
DISCUSSION
The successful use of lytic phages in preventing and treating diverse bacterial infections in humans and animals prompted us to investigate the therapeutic potential of a phage isolate that shows strong lytic activity against P. larvae ERIC I to IV, the causative agents of AFB. At first sight, phage therapy appears to be an ideal alternative to antibiotics, since it represents an autodosing biological pest control method (5, 7). On the basis of experience with therapeutic phages for almost a century and because phages are abundant in natural ecosystems, they are regarded as inherently safe (5). However, rigorous research is required to determine the efficacy of respective therapeutic phages and to confirm their anticipated safety (29). Critical issues that have to be taken into consideration are the following. Phages can convert nontoxic bacteria to toxic strains by lysogenic conversion (9, 30). The pathogenicity of, e.g., Vibrio cholerae, Shiga toxin-producing Escherichia coli, Corynebacterium diphtheriae, and Clostridium botulinum depends on specific prophage-encoded toxins. Moreover, Staphylococcus aureus, Streptococcus pyogenes, and Salmonella enterica serovar Typhimurium harbor a multitude of prophages, and each phage-encoded virulence or fitness factor makes an incremental contribution to the fitness of the lysogen. Other ways in which temperate phages may affect bacterial fitness are by serving as anchor points for genome rearrangements, via gene disruption, by protection from lytic infection, by lysis of competing strains through prophage induction, and via the introduction of new fitness factors (30). In the treatment of AFB, the amplification and application of broad-host-range phages may also have an impact on the gut microbiota of adult honey bees, bee larvae, and the entire microbial ecosystem of the hive. Therefore, to avoid collateral damage by phage therapy, it is important to explore the spectrum of bacteria that are susceptible to the respective therapeutic phage. Finally, for therapeutic success and to avoid resistance, it is also important that the phage selected be sufficiently antagonistic toward the bacteria being targeted.
In the present study, we thoroughly characterized a potential therapeutic phage and developed criteria for safe and efficient phage therapy of AFB. To avoid the potentially hazardous introduction of new infectious agents into the ecosystem by phage therapy, we isolated phages from bee habitats, namely, from the glue-like larval remains of a beehive with clinical symptoms of AFB. Plaque assays were used to select isolates that are highly lytic against P. larvae genotypes ERIC I to IV. Phage HB10c2 was isolated from typical AFB glue-like liquid of a honeycomb, and the natural bacterial host was identified as P. larvae genotype ERIC I by 16S rRNA PCR (20) and by rep-PCR with ERIC primers (17). ERIC I causes the most cases of AFB worldwide (17), whereas ERIC II seems to be restricted to Europe. The former subspecies P. larvae subsp. pulvifaciens, represented by ERIC III and IV, is only rarely isolated.
Purified HB10c2 phages were subjected to transmission electron microscopy. This morphological characterization revealed a long, flexible siphon, a capsid, and a base plate and indicated that phage HB10c2 belongs to the order Caudovirales and the family Siphoviridae with a B2 morphotype.
To obtain a deeper insight into the biology of phage HB10c2, to identify potential risks, and as a basis for the analysis and comparison of newly isolated phages that infect P. larvae, we sequenced and annotated the complete genome of phage HB10c2. Prior to this study, 15 phages of P. larvae were described, and phage HB10c2 exhibits structural similarities to them (31). According to recent developments and further classification of phages at the genomic level in the International Committee on Taxonomy of Viruses, phage HB10c2 could be grouped into the proposed genus “Divavirus.”
Compared to the complete P. larvae genomes sequenced so far (44 to 45% G+C content [32]), the G+C content of the phage genome is lower and similar to that of other Paenibacillus phage genomes (33, 34). Sequence analysis with tRNAscan-SE v.1.21 (35) did not reveal any genes for tRNAs in the genome, which is also consistent with other Paenibacillus phage genomes analyzed. We determined 56 CDS for phage HB10c2, resulting in a coding percentage of 91.3%. Analyses of deduced amino acid sequences revealed that 37 of 56 gene products are similar or identical to gene products of Paenibacillus phage phiIBB_P123 (34). Further similarities to a putative prophage in P. larvae ERIC I strain DSM 25719 could be identified. Whereas P. larvae phage phiIBB_Pl23 harbors a gene for a putative toxin, phage HB10c2 does not have a comparable gene. However, importantly, we identified a gene encoding a putative protein of the beta-lactamase superfamily in phage HB10c2. This is a critical point, since horizontal transfer of antibiotic resistance by phage therapy should be avoided.
BLASTP analysis revealed the conserved domains Terminase_4 (pfam05119) and COG4626, which are responsible for packing DNA into phage capsids. Both proteins are identical to those from Paenibacillus phage phiIBB_Pl23 (34). Terminases are highly conserved, and phylogenetic analyses with other terminases with different known DNA packaging strategies can give first hints about the mechanisms that are used for packaging (36). Recently, Merrill et al. performed a phylogenetic analysis of all of the terminases from all known P. larvae phages (33). The phages that belong to the family Myoviridae seem to use a headful packaging system, whereas siphovirus phiIBB_Pl23 likely has 3′ cohesive ends. As the deduced amino acid of the terminase of HB10c2 is identical to that of Paenibacillus phage phiIBB_Pl23, we presume that phage HB10c2 also contains cos sites and might form concatemers, which was confirmed by pulsed-field gel electrophoresis of genomic DNA of HB10c2 (data not shown).
Most phages lyse their hosts in order to release newly produced phage progeny into the environment. For this, they often use two different kinds of proteins, so-called holins and endolysins. Holins are membrane proteins that assemble in the cytoplasmic membrane and form a pore. The endolysin is a cell wall-degrading enzyme that uses this pore to get into the periplasmic space, where it degrades the peptidoglycan, which leads to bursting of the bacterial cell (37, 38). Downstream of the cluster for structural genes, we identified a lysis cluster consisting of three genes that encode two putative holins and an endolysin. Comparison with other Paenibacillus phages showed that they indeed also harbor either a gene for N-acetylmuramoyl l-alanine amidase, e.g., phage Emery, or a gene for an endo-beta-N-acetylglucosamidase, e.g., phage Davies (33). In particular, endolysins of phages with Gram-positive hosts often act as endopeptidases or amidases and exhibit highly specific activity (39). Therefore, endolysins from Paenibacillus phages could be a further option for the treatment of beehives.
Downstream of these genes, we determined a cluster of genes that might be the lysogeny module that is responsible for integration of the phage genome into the host genome and its further regulation. Among others, we identified genes for an integrase (HB_00029), an excisionase-like protein (HB_00033), and antirepressor proteins (HB_00032 and HB_00037). Therefore, phage HB10c2 is clearly a temperate phage, which makes its suitability for therapeutic use doubtful.
To prevent ecological damage due to the use of HB10c2 as a therapeutic agent against AFB, the bacteriophage was also tested for bacteriolytic activity against environmental and bee-related members of the order Bacillales and members of the larval and adult gut microbiomes of honey bees (28). Bacterial lysis was detected in nine species within the Gram-positive group of Bacillales, mostly in the genus Paenibacillus. The lytic activity of HB10c2 on other bacterial hosts in addition to the pathogen P. larvae is a critical point. On the one hand, this could lead to dysbiosis in the gut of honey bees, the alteration of a healthy microbiota toward a compositional and functional imbalance. On the other hand, the presence of phage HB10c2 in species other than P. larvae could be beneficial, since it may prearm the microbiota against a pathogenic attack. Since the alternative bacterial hosts for phage HB10c2 identified in the gut microbiota are ubiquitous, they may be constantly replaced by bees through food and water intake from the environment. Experimental data to answer the question of whether prophylactic treatment and establishment of a therapeutic phage within a healthy microbial gut community have detrimental or beneficial effects are not available. However, for the development of phage therapy of AFB, this issue seems to be of great importance.
Our breeding assays with P. larvae-infected bee larvae showed no significant effect of therapy with phage HB10c2. This result confirms other analyses that showed that lytic phage activity in liquid or on solid medium is not always predictive of in vivo therapeutic efficacy (11). The reasons for this discrepancy are elusive but may be related to differences in the accessibility of bacterial host cells or the effective phage doses in the respective systems. Moreover, P. larvae is known to sporulate massively in infected bee larvae, which is not the case in the artificial media used (1). P. larvae spores may help bacterial cells to resist lytic phages and by this means could represent a reservoir for perpetual reinfection. This highlights a general problem in the treatment of spore-forming pathogens either by antibiotics or by phage therapy and should be taken into consideration in the future development of AFB therapy.
Conclusion.
This study of P. larvae-specific bacteriophages revealed that rigorous and comprehensive research is required to explore the true potential of phage therapy for treating AFB. This includes the selection of efficient nonlysogenic phages, whole-genome analyses, and representative animal studies that closely simulate the real-life scenario. The lack of efficacy of phage HB10c2 in bee larvae and the potential risk of resistance development in the targeted pathogen suggest that phage cocktails may be required, particularly in preventive applications. From analysis of the phage HB10c2 genome, we conclude that the risk assessment of a putative therapeutic phage should include a data inventory of undesirable virulence and antibiotic resistance genes. Moreover, the host range of the respective therapeutic phage needs to be investigated to ensure that all relevant pathogenic strains are targeted and to minimize dysbiosis of the microbiota and ecological risks. Thus, although it appears relatively simple to isolate phages with activity against the pathogen P. larvae, it seems to be a great challenge to find isolates that meet all efficacy and safety criteria. This may be partly due to the source of the current isolates, since the phages analyzed originated from P. larvae-infected beehives that could not cure themselves. A bird's eye view of the whole genomes of P. larvae and P. larvae phages suggests that phages like HB10c2 contributed to the shaping of the genome of this pathogen. This coevolutionary interaction may have led to a balanced rather than an antagonistic interaction. Accordingly, it may be promising to isolate and characterize phages from other settings or geographical areas.
ACKNOWLEDGMENTS
This work received financial support from the MINAS graduate school and the Niedersächsisches LAVES.
REFERENCES
- 1.Yue D, Nordhoff M, Wieler LH, Genersch E. 2008. Fluorescence in situ hybridization (FISH) analysis of interactions between honeybee larvae and Paenibacillus larvae, the causative agent of American foulbrood of honeybees (Apis mellifera). Environ Microbiol 10:1612–1620. doi: 10.1111/j.1462-2920.2008.01579.x. [DOI] [PubMed] [Google Scholar]
- 2.Genersch E. 2010. American foulbrood in honeybees and its causative agent, Paenibacillus larvae. J Invertebr Pathol 103:S10–S19. doi: 10.1016/j.jip.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 3.Chan QWT, Cornman RS, Briol I, Liao NY, Chan SK, Docking TR, Jackman SD, Taylor GA, Jones SJM, de Graaf DC, Evans JD, Foster LJ. 2011. Updated genome assembly and annotation of Paenibacillus larvae, the agent of American foulbrood disease of honey bees. BMC Genomics 12:450. doi: 10.1186/1471-2164-12-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian B, Fadhil NH, Powell JE, Kwong WK, Moran NA. 2012. Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honeybees. mBio 3:e00377-12. doi: 10.1128/mBio.00377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abedon ST, Kuhl SJ, Blasdel BG, Kutter EM. 2011. Phage treatment of human infections. Bacteriophage 1:66–85. doi: 10.4161/bact.1.2.15845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson RP, Gyles CL, Huff WE, Ojha S, Huff GR, Rath NC, Donoghue AM. 2008. Bacteriophages for prophylaxis and therapy in cattle, poultry and pigs. Anim Health Res Rev 9:201–215. doi: 10.1017/S1466252308001576. [DOI] [PubMed] [Google Scholar]
- 7.Loc-Carrillo C, Abedon ST. 2011. Pros and cons of phage therapy. Bacteriophage 1:111–114. doi: 10.4161/bact.1.2.14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sulakvelidze A, Alavidze Z, Morris JG. 2001. Bacteriophage therapy. Antimicrob Agents Chemother 45:649–659. doi: 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henein A. 2013. What are the limitations on the wider therapeutic use of phage? Bacteriophage 3:e24872. doi: 10.4161/bact.24872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parracho HMRT, Burrowes BH, Enright MC, McConville ML, Harper DR. 2012. The role of regulated clinical trials in the development of bacteriophage therapeutics. J Mol Genet Med 6:279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan KB, Abedon ST, Loc-Carrillo C. 2013. Phage cocktails and the future of phage therapy. Future Microbiol 8:769–783. doi: 10.2217/fmb.13.47. [DOI] [PubMed] [Google Scholar]
- 12.Mazzocco A, Waddell TE, Lingohr E, Johnson RP. 2009. Enumeration of bacteriophages using the small drop plaque assay system. Bacteriophages: methods and protocols, vol. 1 Isolation, characterization and interactions. Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 13.Boulanger P. 2009. Purification of bacteriophages and SDS-PAGE analysis of phage structural proteins from ghost particles. Bacteriophages: methods and protocols, vol. 2 Molecular and applied aspects. Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 14.Wittmann J, Dreiseikelmann B, Rohde M, Meier-Kolthoff JP, Bunk B, Rohde C. 2014. First genome sequences of Achromobacter phages reveal new members of the N4 family. Virol J 11:14. doi: 10.1186/1743-422X-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandrean MA, Barell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 16.Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genersch E, Forsgren E, Pentikäinen AA, Rauch S, Kilwinski J, Fries I. 2006. Reclassification of Paenibacillus larvae subsp. pulvifaciens and Paenibacillus larvae subsp. larvae as Paenibacillus larvae without subspecies differentiation. Int J Syst Evol Microbiol 56:501–511. doi: 10.1099/ijs.0.63928-0. [DOI] [PubMed] [Google Scholar]
- 18.Zwietering MH, Jongenburger I, Rombouts FM, van't Riet K. 1990. Modeling of the bacterial growth curve. Appl Environ Microbiol 56:1875–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aupinel P, Fortini D, Dufour H, Tasei JN, Michaud B, Odoux JF, Pham-Delegue MH. 2005. Improvement of artificial feeding in a standard in vitro method for rearing Apis mellifera larvae. Bull Insectol 58:107–111. [Google Scholar]
- 20.Govan VA, Allsopp MH, Davidson S. 1999. A PCR detection method for rapid identification of Paenibacillus larvae. Appl Environ Microbiol 65:2243–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harland J, Brown SM. 1998. HSV growth, preparation and assay, p 1–4. In Brown SM, MacLean AR (ed), Methods in molecular medicine: herpes simplex virus protocols, vol 10 Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 22.Watson DH, Russell WC, Wildy P. 1963. Electron microscopic particle counts on herpes virus using the phosphotungstate negative staining technique. Virology 19:250–260. doi: 10.1016/0042-6822(63)90062-X. [DOI] [PubMed] [Google Scholar]
- 23.King AMQ, Lefkowith E, Adams MJ, Carstens EB. 2012. Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Elsevier, Oxford, United Kingdom. [Google Scholar]
- 24.Stahly DP, Alippi AM, Bakhiet N, Campana CF, Novak CC, Cox R. 1999. PPL1c, a virulent mutant bacteriophage useful for identification of Paenibacillus larvae subspecies larvae. J Invertebr Pathol 74:295–296. doi: 10.1006/jipa.1999.4893. [DOI] [PubMed] [Google Scholar]
- 25.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 26.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 27.Young I, Wang I, Roof WD. 2000. Phages will out: strategies of host cell lysis. Trends Microbiol 8:120–128. doi: 10.1016/S0966-842X(00)01705-4. [DOI] [PubMed] [Google Scholar]
- 28.Mohr KI, Tebbe CC. 2006. Diversity and phylotype consistency of bacteria in the guts of three bee species (Apoidea) at an oilseed rape field. Environ Microbiol 8:256–272. [DOI] [PubMed] [Google Scholar]
- 29.Pirnay JP, Blasdel BG, Bretau L, Buckling A, Chanishvili N, Clark JR, Corte-Real S, Debarbieux L, Dublanchet A, De Vos D, Gabard J, Garcia M, Goderdzishvill M, Górski A, Hardcastle J, Huys I, Kutter E, Lavigne R, Merabishvili M, Olchawa E, Parikka KJ, Patey O, Pouilot F, Resch G, Rohde C, Scheres J, Skurnik M, Vaneechoutte M, Van Parys L, Verbeken G, Zizi M, Van den Eede G. 2015. Quality and safety requirements for sustainable therapy products. Pharm Res 32:2173–2179. doi: 10.1007/s11095-014-1617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brüssow H, Canchaya C, Hardt WD. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev 68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheflo MA, Gardner AV, Merrill BD, Fisher JN, Lunt BL, Breakwell DP, Grose JH, Burnett SH. 2013. Complete genome sequence of five Paenibacillus larvae bacteriophages. Genome Announc 1:e00668-13. doi: 10.1128/genomeA.00668-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Djukic M, Brzuszkiewicz E, Fünfhaus A, Voss J, Gollnow K, Poppinga L, Liesegang H, Garcia-Gonzalez E, Genersch E, Daniel R. 2014. How to kill the honey bee larva: genomic potential and virulence mechanisms of Paenibacillus larvae. PLoS One 9:e90914. doi: 10.1371/journal.pone.0090914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merrill BD, Grose JH, Breakwell DP, Burnett SH. 2014. Characterization of Paenibacillus larvae bacteriophages and their genomic relationship to firmicute bacteriophages. BMC Genomics 15:745. doi: 10.1186/1471-2164-15-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliveira A, Melo LD, Kropinski AM, Azeredo J. 2013. Complete genome sequence of the broad-host-range Paenibacillus larvae phage phiIBB_Pl23. Genome Announc 1:e00438-13. doi: 10.1128/genomeA.00438-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casjens SR, Gilcrease EB, Winn-Staply DA, Schicklmaier P, Schmieger H, Pedulla ML, Ford ME, Houtz JM, Hatfull GF, Hendrix RW. 2005. The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J Bacteriol 187:1091–1104. doi: 10.1128/JB.187.3.1091-1104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young R. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol Rev 56:430–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young R. 2013. Phage lysis: do we have the hole story yet? Curr Opin Microbiol 16:790–797. doi: 10.1016/j.mib.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wittmann J, Eichenlaub R, Dreiseikelmann B. 2010. The endolysins of bacteriophages CMP1 and CN77 are specific for the lysis of Clavibacter michiganensis strains. Microbiology 156:2366–2373. doi: 10.1099/mic.0.037291-0. [DOI] [PubMed] [Google Scholar]
- 40.Lüken DJ, Janke M, Linau FW, von der Ohe W, Forster R. 2011. Weiterentwicklung einer Methode zur Bienenhaltung unter Laborbedingungen. J Verbr Lebensm 7:141–145. doi: 10.1007/s00003-012-0759-y. [DOI] [Google Scholar]