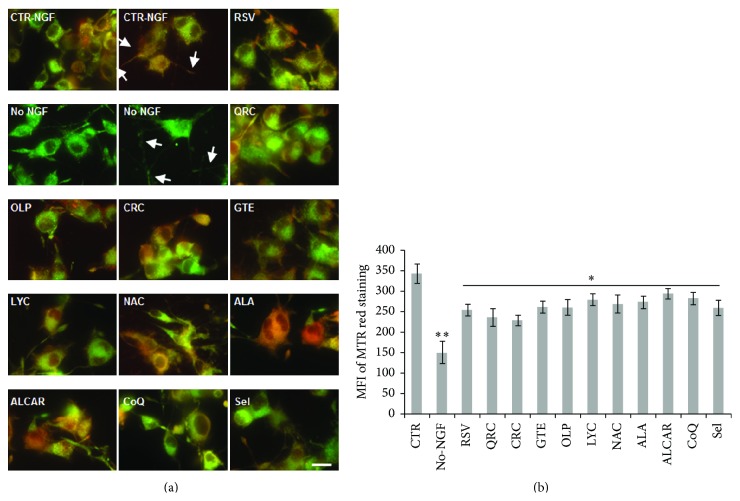
Figure 5.
Analysis of mitochondrial function by MitoTracker Red/Green staining. (a) Representative merged images of neuronal PC12 cells maintained in the presence of NGF (CTR-NGF), NGF-deprived for 12 h (No-NGF), or exposed to NGF-free medium containing the antioxidants. NGF-differentiated PC12 cells were pretreated overnight with of RSV (10 μM), QRC (10 μM), OLP (10 μg/mL), CRC (10 μM), GTE (12.5 μg/mL), LYC (5 μM), NAC (300 μM), ALA (10 μM), ALCAR (10 μM), CoQ (100 nM), or Sel (50 nM) before NGF withdrawal in the presence of the antioxidants. Cells were stained by MitoTracker Red and Green (50 and 200 nM, resp.) during the last 30 min of treatment and observed with a fluorescence microscope (Nikon) equipped with a CCD camera. Images were captured at 60x magnification. Arrowheads point to decreased ΔΨm along neurites. Scale bar = 10 μm. (b) MitoTracker Red fluorescence was quantified by NIH ImageJ, and the MFI was calculated by counting all cells (on average 200 cells) in about 20 random fields for each condition. Data, expressed as MFI, are the mean ± SEM of three experiments in duplicate. ∗ p ≤ 0.05 versus No-NGF, ∗∗ p ≤ 0.01 versus CTR (ANOVA and Dunnett's multiple comparisons test).