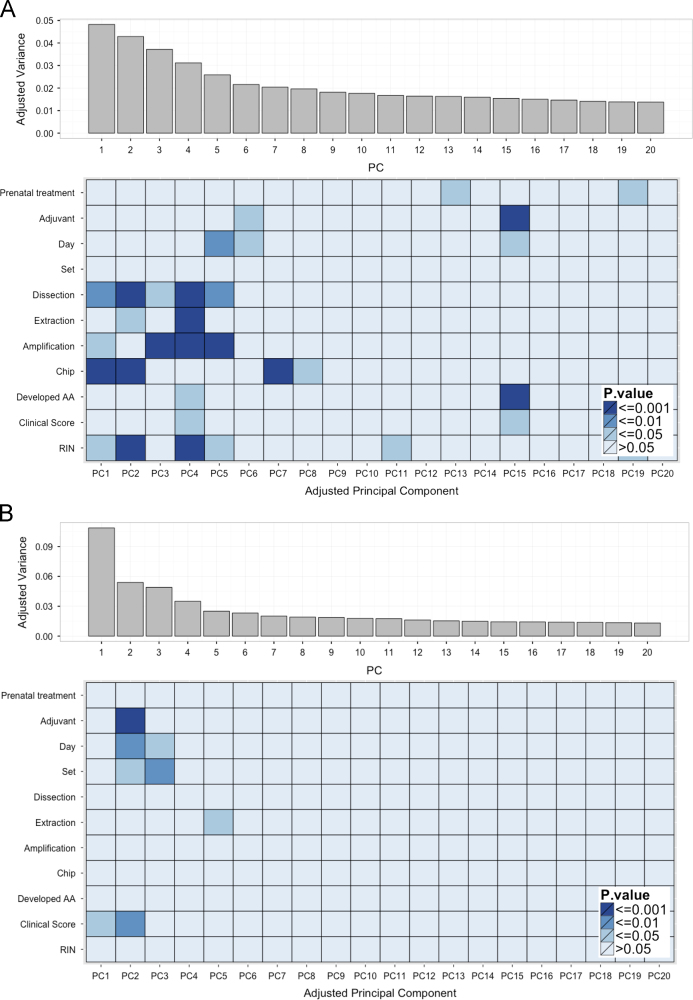
Fig. 6.
Principal component analysis of normalized and filtered datasets from the prefrontal cortex (PFC) and hippocampus (HPC) identified significant batch effects. Adjusted principal components (PC) were analyzed to identify associations with metadata variables and visualize significant sources of variation within the data. (A) In the PFC, batch effects, such dissection, extraction, and amplification round, as well as inter-chip variation, and RNA integrity numbers (RIN) accounted for a large proportion of the adjusted variance. Treatment effects, such as termination day (Day), adjuvant treatment (Adjuvant), total clinical score (Clinical Score), and prenatal treatment (Treatment) accounted for some variance in later principal components. (B) In the HPC, effects from the experimental batch (Set) and extraction round were identified in the first principal components. Effects of termination day, adjuvant exposure, and total clinical score also accounted for a significant proportion of the variance.