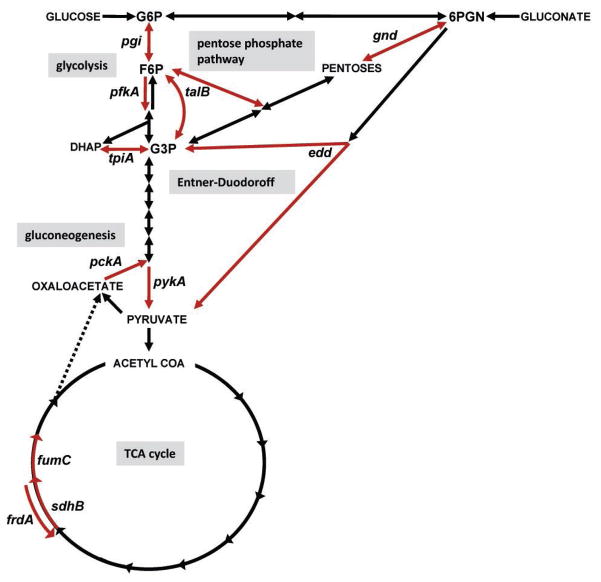
Figure 3. Diagram of central metabolism and map of the specific pathways disrupted by targeted mutations in uropathogenic E. coli.
Carbon sources or biochemical intermediates shared between pathways are indicated in capital letters or abbreviated: G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; G3P, glyceraldehyde-3-phosphate; 6PGN, 6-phosphogluconate. Reactions are denoted with arrows. Specific reactions (red arrows) were targeted by deletion or insertion in E. coli CFT073. In glycolysis: pgi, glucose-6-phosphate isomerase; pfkA, 6-phosphofructokinase transferase; tpiA, triosephosphate isomerase; pykA, pyruvate kinase; in pentose phosphate pathway: gnd, 6-phosphogluconate dehydrogenase; talB, transaldolase; in Entner-Duodoroff pathway: edd, 6-phosphogluconate dehydratase; in gluconeogenesis: pckA, phosphoenolpyruvate carboxykinase; and in the TCA cycle: sdhB, succinate dehydrogenase; fumC, fumarate hydratase; frdA, fumarate reductase. (From, Alteri, Christopher J., Stephanie Himpsl, and Harry L.T. Mobley. 2015. Preferential Use of Central Metabolism in vivo Reveals a Nutritional Basis for Polymicrobial Infection. PLoS Pathogens 11:e1004601.)