Abstract
INTRODUCTION
Genetic data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) has been crucial in advancing the understanding of AD pathophysiology. Here we provide an update on sample collection, scientific progress and opportunities, conceptual issues, and future plans.
METHODS
Lymphoblastoid cell lines and DNA and RNA samples from blood have been collected and banked, and data and biosamples have been widely disseminated. To date, APOE genotyping, genome-wide association study (GWAS), and whole exome and whole genome sequencing (WES, WGS) data have been obtained and disseminated.
RESULTS
ADNI genetic data have been downloaded thousands of times and over 300 publications have resulted, including reports of large scale GWAS by consortia to which ADNI contributed. Many of the first applications of quantitative endophenotype association studies employed ADNI data, including some of the earliest GWAS and pathway-based studies of biospecimen and imaging biomarkers, as well as memory and other clinical/cognitive variables. Other contributions include some of the first WES and WGS data sets and reports in healthy controls, MCI, and AD.
DISCUSSION
Numerous genetic susceptibility and protective markers for AD and disease biomarkers have been identified and replicated using ADNI data, and have heavily implicated immune, mitochondrial, cell cycle/fate, and other biological processes. Early sequencing studies suggest that rare and structural variants are likely to account for significant additional phenotypic variation. Longitudinal analyses of transcriptomic, proteomic, metabolomic, and epigenomic changes will also further elucidate dynamic processes underlying preclinical and prodromal stages of disease. Integration of this unique collection of multi-omics data within a systems biology framework will help to separate truly informative markers of early disease mechanisms and potential novel therapeutic targets from the vast background of less relevant biological processes. Fortunately, a broad swath of the scientific community has accepted this grand challenge.
Keywords: Alzheimer’s Disease Neuroimaging Initiative (ADNI), Alzheimer’s disease (AD), mild cognitive impairment (MCI), genome-wide association studies (GWAS), next generation sequencing (NGS), copy number variation (CNV), biomarkers, magnetic resonance imaging (MRI), positron emission tomography (PET), cerebrospinal fluid (CSF), DNA, RNA, memory, cognition, bioethical issues, precision medicine
1. Introduction
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) was initiated in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies, and nonprofit organizations as a public-private partnership. The first phase (ADNI-1) launched a cohort study of patients with Alzheimer’s disease (AD), mild cognitive impairment (MCI), and cognitively normal older adult controls (CN). A second, American Recovery and Reinvestment Act of 2009 (ARRA) sponsored, “Grand Opportunity” phase (ADNI-GO) followed, and introduced the concept of early mild cognitive impairment (EMCI). ADNI is now completing its third phase (ADNI-2), which also assesses individuals with early or late MCI (LMCI), mild AD, cognitively normal controls, and a new group of older adults with significant memory concerns (SMC) but whose psychometric performance is within normal limits [1], consistent with the recent consensus concept of subjective cognitive decline (SCD) [2]. Detailed information on ADNI is found on the study website (http://www.adni-info.org/). The primary goal of ADNI has been to establish the optimal panel of clinical assessments, magnetic resonance imaging (MRI) and positron emission tomography (PET) imaging measures, as well as other biomarkers from blood and cerebrospinal fluid (CSF), to inform clinical trial design for AD therapeutic development. At the same time, ADNI has been highly productive in generating a wealth of data useful for elucidating disease mechanisms occurring during early stages of preclinical and prodromal AD. For a comprehensive summary of ADNI results see [3] and the other papers in this Special Issue.
Genetics has played an increasingly important role in AD research over the past few decades, starting with the discoveries of rare mutations in APP (amyloid precursor protein), PSEN1 (presenilin-1), and PSEN2 (presenilin-2) that cause early onset autosomal dominant forms of the disease (see [4] for review) and continuing with the association of the common APOE (apolipoprotein E) ε4 allele with sporadic or late onset AD (LOAD). Until recently, only APOE on chromosome 19 had been robustly replicated as a LOAD susceptibility gene. With the advent of large-scale genome-wide association studies (GWAS) conducted by multi-study consortia, a current “top 20” or more list of replicated genes has emerged, which are now undergoing further investigation to identify the biological basis of their risk or protective roles in LOAD [5]. In ADNI, APOE genotype was determined and DNA and cell lines were banked from the beginning of the study. Genome-wide genotyping was also completed, enabling ADNI data to be included in several large and important GWAS studies. Notably, most of these large well-powered meta-analytic GWAS studies are restricted to binary categorical phenotypes, such as clinical diagnosis, and usually only include AD cases and controls.
A relatively unique aspect of ADNI is the systematic longitudinal collection of biomarker data that can serve as quantitative endophenotypes for genetic association studies. In fact, using quantitative endophenotypes as target measures has been found to improve power relative to case-control techniques, as well as to avoid using arbitrary or potentially error prone cut-offs to define case status [6, 7]. Recognizing the value of these quantitative trait loci (QTL) investigations in AD, the ADNI Genetics Core was established at the beginning of the GO phase [8] with the goals of: (1) blood sample processing, genotyping, and dissemination; (2) genome-wide analysis of multidimensional phenotypic data collected on the ADNI cohort; and, (3) serving as a central resource, point of contact, and planning group for genetic studies in ADNI. The first GWAS of an ADNI quantitative phenotype was published in 2009 [9] and progress has been rapid as groups worldwide analyzed the publicly available ADNI genetic data. Shen et al. (2014) provided a detailed review of results from ADNI genetic studies through 2012 [10]. Here, we provide an updated review of these results through 2014, summarize the evolving set of ADNI genetic data, and discuss future directions to maximally leverage this data that we hope will be incorporated in the proposed next phase of ADNI.
The main goal of this update is to describe the genetic data and highlight results generated to date as part of the ADNI study. First, we describe the samples, methods, and basic analysis of the different types of genetic data available in ADNI, including APOE, TOMM40 poly-T repeat, GWAS, whole exome sequences, whole genome sequences, and microarray-based RNA gene expression profiles. This documentation of techniques which have evolved over time should assist the many users of ADNI genetic data to better understand the processing pipelines. Next, we discuss how and the extent to which the various ADNI genetic datasets have been used so far. Third, we highlight novel and important findings generated by relating quantitative phenotypes to the ADNI genetic data. Fourth, we discuss the findings from the ADNI data in the context of a systems biology framework. Next, we describe how ADNI genetic findings could inform drug development, as well as clinical trial design and execution. This is followed by a discussion of the potential for understanding disease biology using induced pluripotent stem cells (iPSC). Next, we describe how ADNI and other genetic data relate to ethical, legal, and social implications (ELSI) of genetic testing, e.g., in returning genetic results on disease risk. Finally, we discuss future directions proposed by the Genetics Core that we hope will be investigated in the proposed next phase of the ADNI study.
2. Materials and Methods
2.1. Sample storage and processing
2.1.1. Sample storage summary
Blood samples are collected at each visit for all participants (i.e., CN, SMC, EMCI, LMCI and AD) according to ADNI protocol for the particular study phase (http://www.adni-info.org/Scientists/ADNIStudyProcedures.aspx). All collected samples are sent to and processed by the NIA-sponsored National Cell Repository for Alzheimer’s Disease (NCRAD; http://ncrad.iu.edu/) according to Standard Operating Procedures briefly summarized below. Baseline blood samples for ADNI-1 were received and processed by the ADNI Biomarker Core with DNA extraction (described below) contributed by Pfizer and performed by Cogenics (now Beckman Coulter Genomics). DNA was then transferred to NCRAD. All samples of genomic material are inventoried and tracked by NCRAD and the ADNI Genetics Core. In addition, collection of PAXgene blood RNA samples (three 2.5-ml tubes per participant) was initiated in the ADNI-GO/2 phases. As of 03/24/2015, there are 777 ADNI-1, 127 ADNI-GO, and 781 ADNI-2 participants with at least one DNA sample from genomic blood stored at NCRAD. 810 ADNI-1, 125 ADNI-GO, and 772 ADNI-2 participants have at least one cell line DNA sample. Because RNA collection was initiated in ADNI-GO, only ADNI-1 participants who continued to ADNI-GO/2 have RNA samples; 290 ADNI-1, 128 ADNI-GO, and 780 ADNI-2 participants have at least one RNA sample stored at NCRAD. Many ADNI-GO/2 participants also have longitudinal RNA collection.
2.1.2. DNA extraction and processing
DNA is extracted from buffy coats using whole blood collected in ethylenediaminetetraacetic acid (EDTA) tubes or buffy coats pre-isolated at the site of collection. The salts of the chelating agent EDTA are used as anticoagulants for genetic testing as they preserve cellular components of blood. Blood samples are centrifuged at 3000 rpm (1500 X g) for 10 minutes at room temperature and the plasma is drawn off and the buffy coat is removed. The buffy coat contains nucleated cells containing DNA. DNA bound to protein is separated using sodium dodecyl sulfate and proteinase K. Then, alcohol is used to precipitate DNA from a high concentration of sodium chloride. Precipitated DNA is collected and transferred into 2-ml or 5-ml tubes following centrifugation and incubation. Once a homogenous solution is obtained and placed inside rotator, DNA concentration is measured on a NanoDrop 1000 Spectrophotometer and DNA tubes are stored in the appropriate −80°C freezer.
2.1.3. RNA extraction and processing
The QIAgen PAXgene Blood RNA Kit (Germantown, MD, USA) is used to purify total RNA from 2.5-ml of human whole blood collected in a PAXgene Blood RNA Tube. The procedure is performed using manual or automated procedures. One 2.5-ml tube is processed to extract RNA and the remaining tubes are stored at −80°C. RNA extraction is performed using the Qiagen QIAcube according to the manufacturer’s protocol. One tube is stored in the 4°C refrigerator before any sample preparation and centrifuged at 3000 X g at room temperature followed by discarding supernatant and mixing RNAse-free water or BR1 resuspension buffer. Samples are then microcentrifuged using QIAcube and eluted samples are transferred to thermal screw cap tubes for incubation for 5 minutes using a benchtop oven pre-heated to 65°C. Quality and quantity of incubated RNA samples is measured and RNA samples are aliquoted in 2-ml screw cap tubes at 2μg/aliquot and stored at −80°C.
2.1.4. Lymphoblastoid cell lines (LCLs)
To establish lymphoblastoid cell lines (LCLs), whole blood samples from the baseline visit are collected in two 8.5-ml tubes coated with acid citrate dextrose. LCLs are created using the white blood cells (WBC) from the buffy coat extracted from whole blood. Cell lines are immortalized by transforming B lymphocytes with the Epstein-Barr virus. WBC are placed in a flask along with a solution that allows permanent cell growth and incubated at 37°C for between 4 weeks and 3 months. The cell-containing solution is then divided and transferred into two larger flasks for further cell growth. Cells are then placed in a vial, along with a preservative, and gradually cooled to freezing temperature to prevent damage to the cell line. The frozen cells are stored in a liquid nitrogen tank at NCRAD.
2.2. Genotyping and sequencing
2.2.1. Apolipoprotein E (APOE) genotyping
APOE genotyping was done using DNA from blood samples from 818 ADNI-1, 128 ADNI-GO, and 778 ADNI-2 participants. For ADNI-1, APOE genotyping was carried out by polymerase chain reaction (PCR) amplification, Hhal restriction enzyme digestion, and subsequent standard gel resolution and visualization processes [11, 12]. For ADNI-GO and ADNI-2 DNA samples, genotyping was performed by Prevention Genetics (Marshfield, WI, USA) and LGC Genomics (Beverly, MA, USA). Prevention Genetics employed array processing using allele-specific PCR with universal molecular beacons [13, 14]. At LGC, assays were performed using competitive allele-specific PCR, enabling bi-allelic scoring of single nucleotide polymorphisms (SNPs). Assay kits were added to DNA samples, followed by thermal cycling reaction and an end-point fluorescent read. Genotypes were called using LGC Genomics’ in-house Kraken software (http://www.lgcgroup.com/products/genotyping-software/kraken/#.VRCH6I7F98E) and were returned to the ADNI Genetics Core after manual quality control (QC). All APOE genotype data underwent further QC checks, including sex and identity checks, and potential problems were identified and corrected through communication with NCRAD and other cores. Final quality-controlled data were posted on the ADNI LONI website (http://adni.loni.usc.edu).
2.2.2. Translocase of outer mitochondrial membrane 40 homolog (TOMM40) poly-T repeat genotyping
The TOMM40 poly-T repeat was assessed in the ADNI-1 cohort because an association had been previously reported between the TOMM40 poly-T repeat and AD [15]. Genotyping was performed by Polymorphic DNA Technologies, Inc. (Alameda, CA, USA) for 757 individuals from ADNI-1, with support for the assays provided by Allen Roses (Duke University). Details of the poly-T assay and bioinformatics analyses using long-range PCR and DNA sequencing are available in [15].
2.2.3. Genome-wide array genotyping
Genome-wide genotyping was performed using the Illumina Human 610-Quad BeadChip (Illumina, Inc., San Diego, CA, USA) for ADNI-1 individuals by TGen (Phoenix, AZ, USA) and using the OmniExpress BeadChip for ADNI-GO/2 individuals by the Center for Applied Genomics of Children’s Hospital of Philadelphia (Philadelphia, PA, USA). Detailed genotyping protocols were described previously [8]. All assays were performed according to manufacturer protocols. Bead intensity data were used to call genotypes using BeadStudio 3.2 (Illumina) for the first release of ADNI-1 data, GenomeStudio v2009.1 (Illumina) for the second release of ADNI-1 data, and GenomeStudio v2011.1 (Illumina) for ADNI-GO/2. All genotype data was quality controlled, including checks for sex and identity, and the quality-controlled data were released on the ADNI LONI website (http://adni.loni.usc.edu) in Final Report and/or PLINK data formats. Of note, samples from the 818 individuals in the whole genome sequencing (WGS) sub-study described below (Section 2.2.5) underwent genome-wide genotyping using the Illumina Omni 2.5M BeadChip performed by Illumina. Thus, multiple genome-wide genotype array datasets are available for some ADNI participants. After QC, the Omni 2.5M genotype data in PLINK format were posted on the LONI website (http://adni.loni.usc.edu) in February 2014.
2.2.4. Whole Exome Sequencing (WES)
Whole exome sequencing (WES) of 18 ADNI-1 participants with a diagnosis of MCI at baseline was performed on blood-derived genomic DNA samples at TGen (www.tgen.org/). Participants were selected for a small extreme-trait design. Specifically, nine age- and education-matched pairs of non-Hispanic Caucasian, male, right-handed participants with amnestic MCI were selected on the basis of rapid versus slow hippocampal volume change on MRI over 2 years (nine participants with rapid rates of atrophy and nine with slow rates of atrophy) [16, 17]. All participants had APOE ε3/ε3 genotype, as one goal of this small study was to identify coding variants other than the APOE ε4 allele that are associated with rate of hippocampal volume loss. Sequences were enriched through hybridization using the Agilent’s SureSelect Human All Exon 50Mb kit following the manufacture’s protocol (www.genomics.agilent.com/). The Agilent kit captured an exome that was approximately 50Mb in size, covering ~21,000 genes. These samples could then be sequenced together on one lane of the flowcell and segregated later for analysis using their molecular bar-codes as tags. Samples were sequenced across multiple flowcell lanes to account for any possible lane effects. These libraries were sequenced on the Illumina HiSeq2000 using paired-end read chemistry and read lengths of 105bp. Each sample was sequenced two times and the resulting fastq files were posted to the ADNI LONI website (http://adni.loni.usc.edu) after QC.
2.2.5. Whole Genome Sequencing (WGS)
Whole genome sequencing (WGS) was performed on blood-derived genomic DNA samples from 818 ADNI-1/GO/2 participants with support from the Brin-Wojcicki Foundation and the Alzheimer’s Association. This research sequencing was performed in a non-CLIA (see Section 4.4) laboratory at Illumina. 810 of the ADNI WGS datasets were released in August 2013. Samples were sequenced on the Illumina HiSeq2000 using paired-end read chemistry and read-length of 100bp at 30–40X coverage. The resulting BAM and VCF files generated by Illumina using the CASAVA software were archived at three physical locations, including LONI at the University of Southern California, Indiana University, and the Broad Institute. QC steps included participant sex check, participant identity check, and variant quality check of the Illumina-generated VCF files. The participant sex check was conducted by comparing sex estimated using Illumina Omni2.5M GWAS data and self-reported sex information from the ADNI database. Participant identity was cross-checked by determining: (1) concordance of a subset of fingerprint SNPs between prior genotype data and Omni2.5M genotype data; and, (2) concordance of all quality-controlled SNPs between Omni2.5M genotype data and WGS VCF data. The quality of variants passing sequencing QC in the WGS VCF files was assessed by comparing with Omni2.5M genotype data. Since Illumina’s CASAVA software calls variants on a single sample only, the Broad Institute converted the BAM files into original fastq files, a text-based format for storing both sequence reads and their corresponding quality information in Phred format, and re-aligned and re-called the variants using Broad GATK. Briefly, short-read sequences were mapped to the reference human genome using BWA-mem and potential PCR duplicates were removed. After completing initial alignment, the alignment was further refined by locally realigning any suspicious reads. The reported base calling quality scores obtained from the sequencer were recalibrated to account for covariates of base errors such as sequencing technology and machine cycle. Finally, the realigned reads were written to a BAM file for further analysis. The analysis-ready BAM files were assessed to identify all variants with statistical evidence for an alternate allele present among samples using GATK HaplotypeCaller for multi-sample variant callings. Variants that passed recommended variation quality criteria were assessed by comparing with the Illumina Omni 2.5M genotyping data to estimate the concordance rate for each individual. Finally, the resulting VCF files were posted to the ADNI LONI website (http://adni.loni.usc.edu).
2.3. Blood RNA Expression Microarray Profiling
Gene expression or RNA profiling is a new type of data for ADNI that has long been planned but was not previously supported by existing funding. Fortunately RNA profiling on selected baseline blood samples was contributed by Bristol-Myers Squibb (BMS) and performed at the BMS laboratories for 811 ADNI participants included in the WGS sub-study. The Affymetrix Human Genome U219 Array (Affymetrix, Santa Clara, CA) was used for expression profiling. Peripheral blood samples were collected using PAXgene tubes for RNA analysis. Total RNA was extracted using the PAXgene Blood RNA Kit, following the protocol provided by the manufacturer. The quantity and quality of extracted RNA were assessed using the NanoDrop and the Agilent Bioanalyzer, respectively. Blood RNA samples from 64 participants did not pass QC and were excluded. Samples were randomized to plates, with checks to ensure sex and diagnosis balance, and hybridized to Affymetrix Human Genome U219 array plate. Array hybridization, washing, staining, and scanning were carried out in an Affymetrix GeneTitan system. The quality of gene expression data, including sample quality and hybridization and overall signal quality, was analyzed using Affymetrix Expression Console software and Partek Genomic Suite 6.6, according to standard QC criteria provided by each software package. Raw expression values obtained directly from CEL files were pre-processed using the Robust Multi-chip Average (RMA) normalization method [18]. The Affymetrix HG U219 Array contains 530,467 probes for 49,293 transcripts. All Affymetrix U219 probe sets were mapped and annotated with reference to the human genome (hg19). The Genetics Core performed several additional QC steps using the RMA normalized expression array data. First, the sex of samples was checked using sex-specific gene expression data, including XIST and USP9Y [19]. Second, sample identity was verified on the basis of expression profiling to Omni2.5M genotype match using a Bayesian method to predict individual SNP genotypes from gene expression data [20]. Briefly, the 1,000 most significant SNP-transcript cis-eQTL pairs from quality-controlled gene expression and genotype data were used to estimate the posterior probability for a match between gene expression and genotype data. The quality-controlled gene expression profiles were completed and released to the ADNI LONI website (http://adni.loni.usc.edu) in late April 2015.
2.4. Metabolomic and Lipidomics Profiling
Another new and important data type for ADNI expected to become available in mid-2015 is metabolomics profiling. As a brief background, the metabolome is an organism’s repertoire of metabolites or small molecules present in cells, tissues, body fluids, and organ systems. Metabolomics is the comprehensive study of metabolites and metabolism or biochemical processes at the global or “-omics” level. In contrast to the RNA profiling discussed above that reflects transcription processing and gene products, metabolic profiling assays the end products of cellular processes and can provide a more complete picture of cellular physiology and pathophysiology. This is a relatively new and rapidly expanding field that is recognized as having the potential to significantly impact medical practice [21–24].
Over the past few years, the ADNI Biomarker and Genetics Cores have collaborated with Dr. Rima Kaddurah-Daouk (Duke University) and colleagues to initiate metabolomics profiling of ADNI samples. Several initial studies were conducted using samples from Duke and the University of Pennsylvania [25–27]. Currently, ADNI-1 plasma samples are being profiled, as are additional samples from the Indiana University Memory and Aging Study (IMAS) [28]. Profiling is expected to include broad biochemical coverage with over 500 metabolites measured in each participant to permit analysis of metabolic changes within multiple pathways and networks. A biochemical database will be created and released on the ADNI LONI website (http://adni.loni.usc.edu). We expect that baseline data from ADNI-1 will be profiled and released by end of 2015.
3. Results
3.1. Data use statistics
Since the initial GWAS genotyping of ADNI-1 samples by TGen, there were 31 sample requests fulfilled by NCRAD. As of 3/24/2015, 8333 ADNI DNA samples were used for APOE and GWAS genotyping, TOMM40 Poly-T genotyping, WES, WGS, replication of genetic findings from other cohorts, and other studies. 811 RNA PaxGene tubes were used for the RNA expression profiling by BMS. ADNI genetic data dissemination is managed and tracked by the Informatics Core based at LONI. From 2009 through the end of 2014, ADNI GWAS and/or other genetic data were downloaded approximately 107,000 times. For WGS, 12 sets of the extensive (~150 TB) WGS raw data set (BAM files) have been disseminated by hard drive array, which were return shipped to requestors.
3.2. Publication Statistics
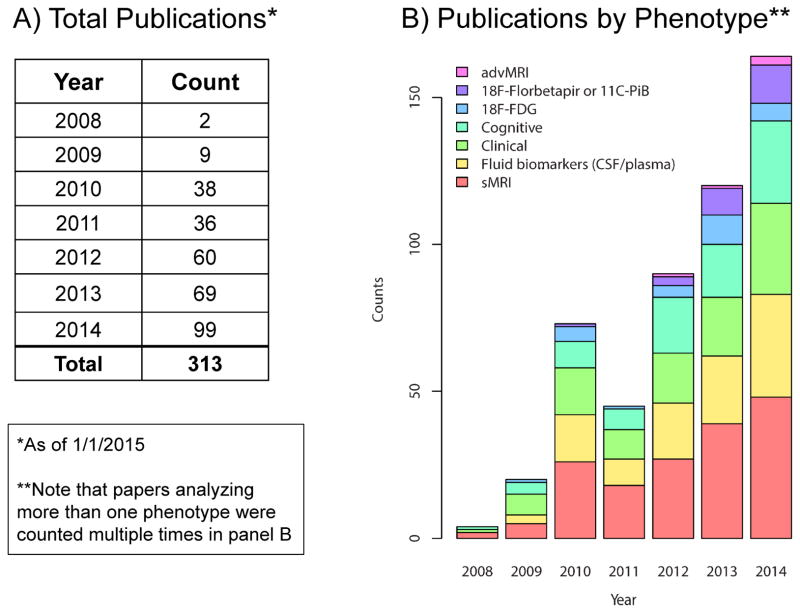
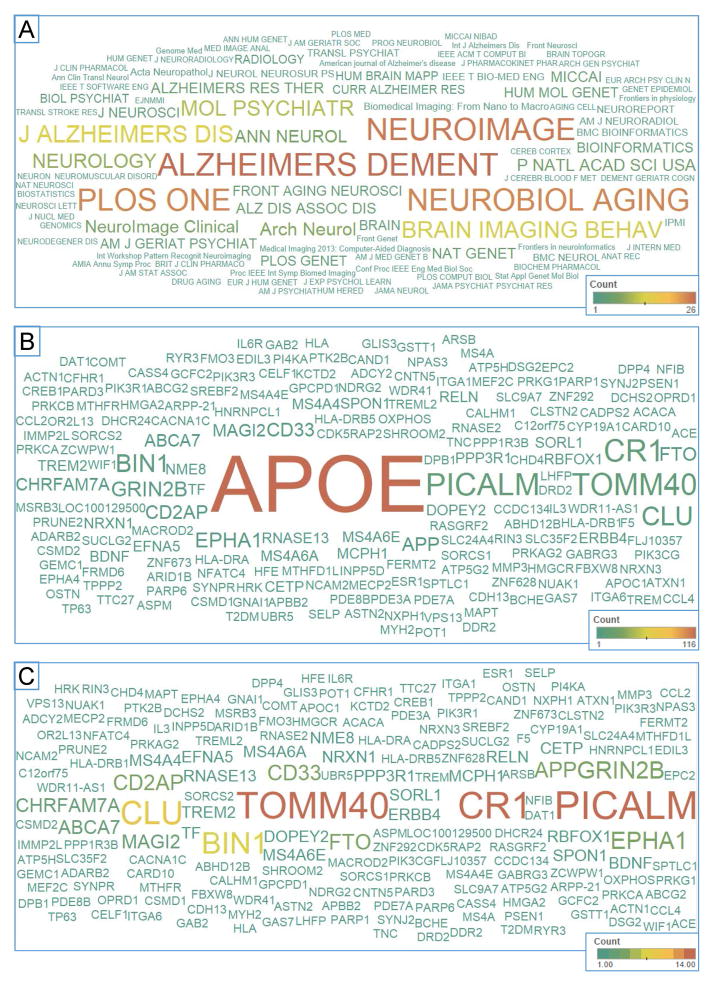
Here we briefly summarize the ADNI genetic studies published up to 1/1/2015, where ADNI APOE, GWAS, or sequencing data were used. We searched the PubMed database using the EndNote X7 online search tool with the following three criteria: (1) the “All Fields” contain “Alzheimer’s Disease Neuroimaging Initiative”; (2) the “All Fields” contain “APOE”, “apolipoprotein”, “gene”, “genetic”, “genetics”, “genome”, “genomic”, or “genomics”; and, (3) the “Year” field value is between 2008 and 2014. We integrated the search results with the ADNI publication database maintained by the ADNI Data and Publications Committee. We manually reviewed all the abstracts and identified 313 relevant ADNI genetic publications through this extensive search. These publications include 2 in 2008, 9 in 2009, 38 in 2010, 36 in 2011, 60 in 2012, 69 in 2013, and 99 in 2014; see Supplementary Material (ADNI_Genetics_papers_2008-2014.enlx). In these publications, ADNI genetic data were studied together with one or more of the ADNI multi-dimensional phenotypic data sets, including clinical information, structural MRI (sMRI), advanced MRI such as diffusion tensor imaging (DTI), resting state fMRI, or arterial spin labeled (ASL) perfusion MRI, [18F]Florbetapir (AV-45) or [11C]Pittsburgh Compound B (PiB) PET, [18F]Fluorodeoxyglucose (FDG) PET, CSF or plasma biomarkers, and/or cognitive measures. Figure 1 shows the distribution of ADNI genetic publications grouped by assessed phenotype(s) from 2008 to 2014. Note that papers analyzing more than one phenotype were counted multiple times. Figure 2a shows the word cloud of journal names where these papers were published, where the color and size of a journal name correspond to the number of papers published in that journal. Figures 2b and 2c show the word cloud of gene names appearing in the paper abstracts, where the color and size of a gene name correspond to the number of abstracts mentioning the gene. Note that Figures 2b and 2c represent the results with and without APOE included, respectively.
Figure 1.
ADNI genetic data usage and reports (2008–2014)
(A) Total publications by year and (B) by phenotype category using ADNI genetic data are displayed. Note that papers analyzing more than one phenotype were counted multiple times in panel B. (advMRI = studies using advanced MRI techniques (diffusion tensor imaging, resting-state functional MRI, arterial spin labeling perfusion MRI); 18F-Florbetapir or 11C-PiB = studies using [18F]Florbetapir or [11C]PiB; 18F-FDG = [18F]FDG studies; Cognitive = studies utilizing neuropsychological test performance data; Clinical = studies using clinical data, such as diagnosis; Fluid biomarkers (CSF/plasma) = studies using CSF or plasma-based fluid biomarkers; sMRI = structural MRI studies)
Figure 2.
Common journals and reported genes in manuscripts using ADNI genetic data
(A) A word cloud of journal names where papers utilizing ADNI genetic data were published is shown with the color and size of a journal name corresponding to the number of papers published in that journal. Word clouds of gene names appears in these paper abstracts, (B) with and (C) without including APOE, are displayed with the color and size of a gene name corresponding to the number of abstracts mentioning the gene.
3.3. Genetic Association Results by Phenotype
Since the first ADNI GWAS publication by Potkin et al. [9], numerous genetic analyses have used the ADNI data alone or in combination with other cohorts. The papers published between 2009 and 2012 were summarized in detail in [10]. Publications including ADNI genetic data continued to increase rapidly in 2013 and 2014. As shown in Figure 1, different types of qualitative and quantitative phenotypes have been studied with genetic data in ADNI. Below, we summarize and selectively review recent findings grouped by phenotypic category.
3.3.1. Clinical Diagnosis (Case vs. Control)
One of the most prominent case control studies in LOAD was conducted by Lambert et al. [5], who performed a meta-analysis of 74,046 individuals, including the ADNI cohort, and identified 11 new susceptibility loci for LOAD. Boada et al. [29] performed another multi-cohort study, including ADNI, and identified a novel LOAD association at the adenosine triphosphate (ATP) synthase, H+ transporting, mitochondrial F0 (ATP5H)/Potassium channel tetramerization domain-containing protein 2 (KCTD2) locus. The ATP5H/KCTD2 locus has been related to mitochondrial energy production and neuronal hyperpolarization during cellular stress conditions. Leduc et al. [30] performed a three-cohort study, including ADNI, and identified rs3846662 in the 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) gene as a potent genetic modifier for AD risk, age of onset, and MCI to AD conversion. This finding is noteworthy since the use of statins (HMGCR inhibitors) during mid-life has been demonstrated to reduce the risk of developing sporadic AD by as much as 70%, although no AD-related benefits of statins have been shown in clinical AD patients. Biffi et al. [31] performed a targeted genetic association study in ADNI and identified variations in the oxidative phosphorylation gene-set associated with AD status and conversion from CN to MCI. Kim et al. [32] performed a gene-based analysis of relatively rare alleles (minor allele frequency≤0.03) in ADNI, coupled with several other AD cohorts from dbGaP, and identified an association between ZNF628 (zinc finger protein 628; a transcription factor coding gene) and AD. Finally, Desikan et al. reported on the genetic overlap between AD and Parkinson’s disease (PD) based on large GWAS data sets including ADNI (total n=89,904) and identified a marker in the extended MAPT (microtubule-associated protein tau) region (A allele of rs393152) associated with both AD and PD [33]. Comparisons of common and divergent pathways in neurodegenerative diseases including AD, PD, amyotrophic lateral sclerosis, and other conditions appears to be a promising strategy as discussed in [34].
3.3.2. Structural MRI
Data extracted from structural MRI scans have been the most widely analyzed phenotypes in ADNI genetic studies as was described in Shen et al. [10]. As an example, Nho et al. [16, 17], using an extreme phenotype design discussed above in Section 2.2.4, identified genetic variations within PARP1 (poly (ADP-ribose) polymerase 1) and CARD10 (caspase recruitment domain family, member 10) genes associated with a more rapid rate of hippocampal volume loss in MCI. Recently, the same extreme phenotype imaging genetics design was used to identify REST (RE1-silencing transcription factor) as a protective variant [35, 36], which was consistent with an elegant independent series of analyses in post-mortem tissue and cellular models reported by Lu and colleagues in Nature [37]. Hohman et al. [38, 39] performed two ADNI studies to examine genetic modification of the relationship between other AD biomarkers and MRI-based neurodegeneration. In one study [38], they identified rs4728029 from the POT1 (protection of telomeres 1) gene, which modifies the relationship between phosphorylated tau and ventricular dilation. In the other study [39], they identified several SNPs that modify the relationships between amyloid-beta (Aβ) or tau positivity and neurodegeneration. Roussotte et al. performed a series of ADNI MRI-based genetic studies [40–43] and identified a common variant in the DAT1 (dopamine transporter; also known as SLC6A3, solute carrier family 6 (neurotransmitter transporter), member 3) gene associated with faster ventricular expansion [40], combined effects of LOAD risk variants in the CLU (clusterin) and APOE genes on ventricular expansion [41], a common variant in the OPRD1 (opioid receptor, delta 1) gene associated with smaller regional brain volumes [42], and a variant in the RASGRF2 (Ras protein-specific guanine nucleotide-releasing factor 2) gene associated with larger cortical volume but faster longitudinal ventricular expansion [43]. Luis et al. [44] performed an MRI-based genetic study on the TREM2 (triggering receptor expressed on myeloid cells 2) AD risk variant (rs75932628), which included ADNI and a Spanish cohort, and identified an association between the TREM2 variant and frontobasal grey matter loss. Rajagopalan et al. also found that carriers of the risk-conferring variant in TREM2 had faster atrophy in the temporal lobes compared to non-carriers [45]. Koran et al. [46] evaluated the effect of genetic interactions within inositol-related pathways on longitudinal MRI measures of the inferior lateral ventricles and identified several genetic interactions associated with longitudinal changes in ventricle size. Andrawis et al. [47] demonstrated that APOE had a significant effect on longitudinal change in the hippocampus, with greater change in MCI and AD participants who were APOE ε4 positive. Finally, Apostolova et al. [48] demonstrated that APOE genotype alters the timing and sensitivity of hippocampal volume and CSF biomarkers for predicting MCI to AD conversion, with better prediction using hippocampal volume in APOE ε4 positive MCI participants and better prediction using p-tau in APOE ε4 negative participants.
3.3.3. Advanced MRI
ADNI-GO/2 introduced several advanced MRI modalities, including DTI, resting-state fMRI, and ASL perfusion MRI. In each case these advanced techniques were limited to sites employing a particular scanner vendor effectively reducing the sample to about one third of the cohort for each scan type and limiting the power for genetic analyses (see Jack et al. MRI Core report in this Special Issue). Given the modest sample size of these data sets, only a few genetic studies have been performed to date, using only the ADNI DTI data. Notably, however, combining data from two cohorts, Jahanshad et al. [49] performed the first connectome-wide and genome-wide association study and identified an association between a variant in SPON1 (spondin 1; rs2618516) and brain anatomical fiber connectivity. Warstadt et al. [50] fitted a multi-locus genetic model within white matter areas associated with serum cholesterol and identified that CETP (cholesteryl ester transfer protein, plasma; increased rs5882 G-allele dosage) was associated with higher fractional anisotropy and lower radial and mean diffusivity on DTI. Several publications also proposed new methods to boost the power to detect genetic associations with diffusion imaging measures in ADNI, by using bivariate genetic analysis of MRI and DTI and an approach called “seemingly unrelated regression” which integrates genetic information from different imaging biomarkers [51–53].
3.3.4. Amyloid PET ([11C]PiB; [18F]Florbetapir)
An early targeted study using a modest sized ADNI-1 [11C]PiB PET sample examined the amyloid pathway [54]. The number of genetic studies with amyloid PET phenotypes grew rapidly in 2013 and 2014. For example, Bradshaw et al. [55] examined the CD33 (CD33 molecule) LOAD susceptibility locus (rs3865444) with the ADNI-1 [11C]PiB PET data and identified an association with increased [11C]PiB binding. Ramanan et al. [56] performed the first GWAS with amyloid PET as the phenotype using the ADNI-GO/2 [18F]Florbetapir scan data and reported that APOE and BCHE (butyrylcholinesterase) were independent modulators of cerebral amyloid deposition, together accounting for nearly 15% of the variance in cross-sectional amyloid burden. Thambisetty et al. [57] examined the CR1 (complement component (3b/4b) receptor 1) LOAD susceptibility locus (rs3818361) using ADNI-1 [11C]PiB PET data, combined with [11C]PiB PET data from sub-studies of the Baltimore Longitudinal Study of Aging (BLSA), and reported that risk allele carriers had lower brain amyloid burden compared to non-carriers. Swaminathan et al. [58] examined the association between plasma Aβ from peripheral blood and cortical amyloid deposition on [11C]PiB PET and found that this relationship was modulated by APOE ε4 genotype. Murphy et al. [59] mapped the effects of APOE ε4 on ADNI [18F]Florbetapir scans and found a significant effect in the four cortical regions examined. Risacher et al. [60] investigated the role of APOE genotype in EMCI and found that the APOE ε4 allele was associated with increased amyloid accumulation on [18F]Florbetapir PET, lower CSF Aβ1-42, and increased CSF tau levels in EMCI and CN. Recently, Risacher et al. [1] also examined the role of APOE in an expanded sample that included the new SMC group introduced in ADNI-2. SMC participants who were carriers of at least one APOE ε4 allele showed greater amyloid deposition on [18F]Florbetapir PET, lower CSF Aβ1-42, and increased CSF tau levels compared to those who were APOE ε4 non-carriers. Interestingly, an effect of APOE ε4 was not observed for hypometabolism or medial temporal lobe atrophy in the SMC group consistent with the concept that those markers may be downstream of the amyloid and tau proteinopathies detected by PET and in CSF [61]. Finally, several amyloid PET studies examined various facets of epistasis or gene-gene interactions. Yan et al. [62] performed a transcriptome-guided amyloid imaging genetic analysis and identified bi-multivariate associations between APOE SNPs and brain-wide amyloid imaging measures. Hohman et al. [63] examined epistatic relationships between genes involved in amyloid and tau pathophysiology using [18F]Florbetapir PET imaging as the target phenotype. This study identified three significant interactions between a SNP in GSK3β (glycogen synthase kinase 3 beta; rs334543) and SNPs in each of the following genes: APBB2 (amyloid beta (A4) precursor protein-binding, family B, member 2; rs2585590, rs3098914) and APP (amyloid beta (A4) precursor protein; rs457581).
3.3.5. [18F]Fluorodeoxyglucose (FDG) PET
Relatively few recent reports specifically examined genetic associations with [18F]FDG PET as an isolated phenotype. However, several studies performed integrative analyses of genetics, [18F]FDG PET, and other imaging and biomarker data for predicting outcomes of interest. For example, Schraml et al. [64] proposed a hypometabolic convergence index (HCI) to provide a summarized measure of hypometabolism on [18F]FDG PET and identified a significant association between HCI and APOE ε4 dose using ADNI data. [18F]FDG PET measures have also been analyzed with genetic data such as APOE ε4 status, along with other multidimensional biomarkers, to classify AD status [65], predict cognitive decline [66], or predict MCI to AD conversion [67].
3.3.6. Fluid Biomarkers (CSF and Plasma)
Fluid biomarkers are another frequently analyzed category of quantitative phenotypes used in ADNI genetic studies. Kim et al. [28] examined the influence of genetic variation on plasma protein levels using the multi-analyte Rules Based Medicine (RBM) panel and identified multiple novel associations. For example, the CFHR1 (complement factor H-related 1; rs7517126) and CFH (complement factor H; rs6677604) genes were discovered to be very highly associated with plasma complement factor H-related protein 1 (CFHR1) level. This study also confirmed previously identified gene-protein associations for the interleukin-6 receptor (IL6R), chemokine C-C motif (CC) ligand 4, angiotensin-converting enzyme (ACE), and angiotensinogen. Cruchaga et al. [68] performed a GWAS of CSF tau/phosphorylated tau (pTau181) levels and identified three loci: one between GEMC1 (geminin coiled-coil domain containing; also known as GMNC) and OSTN (osteocrin), one within GLIS3 (GLIS family zinc finger 3), and one within the TREM gene cluster. Chouraki et al. [69] performed a genome-wide meta-analysis of plasma Aβ peptide levels and found a significant association between the CTXN3 (cortexin 3) gene and plasma Aβ1-42 levels. Kauwe et al. [70] performed a GWAS of CSF levels of 59 AD candidate proteins and identified genetic associations with CSF levels of 5 proteins, including ACE, chemokine CC ligand 2, chemokine CC ligand 4, IL6R, and matrix metalloproteinase-3 (MMP-3). As mentioned earlier, Hohman et al. performed two ADNI studies [38, 39] to examine genetic modification of relationship between CSF biomarkers and MRI-based neurodegeneration and identified rs4728029 from the POT1 gene that modifies the relationship between phosphorylated tau and ventricular dilation [38] and several SNPs that modify the relationships between amyloid or tau positivity and neurodegeneration [39]. Ramirez et al. [71] performed a GWAS of CSF Aβ1-42 and pTau181 levels and found an association between Aβ1-42 level and rs62256378 in SUCLG2 (succinate-CoA ligase, GDP-forming, beta subunit), as well as an interaction between APOE ε4 genotype and this SNP. In the above-mentioned study by Risacher and colleagues [1], individuals with SMC who were APOE ε4 carriers showed lower Aβ1-42 and higher tau and pTau181 than SMC APOE ε4 non-carriers. A recent cross-sectional and longitudinal analysis [72] of the relationship between CSF and PET amyloid biomarkers by Toledo et al. [72] found that APOE modified the relationship between these AD biomarkers and suggested that they measure different aspects of Aβ pathology in AD. Finally, as mentioned above, Apostolova et al. [48] found that p-tau was the best predictor for MCI to AD conversion in APOE ε4 negative participants, while hippocampal volume was the best predictor in APOE ε4 positive MCI participants.
3.3.7. Cognitive Performance
Studies including ADNI data have established that the effect of APOE on cognitive performance is more complex than a simple extension of its association with AD risk [60, 73–76]. For example, one study demonstrated an interactive effect of APOE and amyloid pathology on cognitive decline, characterized by APOE ε4 carriers with high amyloid burden exhibiting greater cognitive decline [76]. This finding was consistent with an earlier analysis in the Mayo Clinic Study of Aging that suggested that APOE isoforms modify the association between amyloid load and cognition [77].
Other genetic analyses of cognition have expanded the scope beyond APOE to study other known AD risk loci [74, 78], as well as to discover novel associations. Sherva et al. reported that the SPON1 gene was associated with longitudinal global cognitive decline [79]. This SPON1 finding is notable in view of the above-mentioned report of an association of SPON1 with cerebral connectivity in both ADNI and a young twin cohort [49]. Ramanan et al. identified an association between the pro-apoptotic gene FASTKD2 (fas-associated serine/threonine kinase domains 2) and cross-sectional episodic memory performance [80, 81]. The Ramanan et al. [80, 81] memory GWAS is noteworthy for its large sample consisting of data from the United States Health and Retirement Study (HRS), ADNI, AddNeuroMed, the Religious Order Study (ROS), the Rush Memory and Aging Project (MAP), and the Indiana Memory and Aging Study (IMAS) yielding a total discovery and replication sample of 14,781 individuals. The novel SNP discovered in this study exhibited a protective effect on memory performance and hippocampal structure (volume and grey matter density on structural MRI) and was associated with decreased CSF levels of apoptotic mediators, consistent with a neuroprotective effect of this gene variant. In related work, the ADNI dataset contributed to the large scale genome-wide screens of brain measures conducted by the ENIGMA Consortium [82]. In one analysis of GWAS data and MRI measures from over 50 cohorts worldwide, the ENIGMA consortium, in partnership with ADNI, discovered 8 common genetic variants associated with the volume of subcortical brain structures, including the hippocampus; several of the implicated genes are associated in aggregate with disease risk and have known roles in cell guidance, apoptosis, and other physiological processes.
Other studies have related genes to cognition following the discovery of a primary association with a non-cognitive phenotype, including ASTND2 (astrotactin 2) and GRIN2B (glutamate receptor, ionotropic, N-methyl D-aspartate 2B) with hippocampal and temporal lobe atrophy on MRI [83, 84], PSEN1 (presenilin 1) with CSF amyloid levels [85], and DAT1 with AD case-control status [40]. Alternatively, pathway analysis has been used to elucidate collective effects of multiple genes on memory [86], executive functioning [87], and depressive symptoms [88]. Notably, pathway analysis of memory in the ADNI-1 sample (N=742) [86] displayed substantial overlap of underlying mechanisms (particularly of pathways related to cholinergic neurotransmission, cell adhesion, and inflammation) with a subsequent pathway analysis of memory in the population-based HRS sample (N=6,705) [80], highlighting the potential utility of pathway-based approaches [34, 89] to yield replicable associations from otherwise diverse studies.
Overall, these findings provide a foundation for meeting the broader ADNI aim of enhancing clinical trial design and interpretation. Cognitive phenotypes are thought to have substantial heritability (up to 80%) based on twin studies, but a substantial portion of this heritability is still uncharacterized, and known genes appear to exhibit modest individual effect sizes [90–92]. Although APOE has shown the most consistent relationship with cognition in AD and non-AD settings, its effects depend on interactions with other factors such as age [73, 93, 94] and biomarker status [60, 75], so accounting for these interactions might facilitate risk enrichment and stratification in clinical trials [95]. The use of genetic profiling for risk stratification to enhance clinical trial design is discussed in greater detail in Section 4.2 below. Other heavily-investigated genes, such as BDNF (brain-derived neurotrophic factor) [96–98] and KIBRA (WWC1; WW and C2 domain containing 1) [99–102], have displayed inconsistent relationships with cognitive outcomes, potentially due to the complexity of the cognitive “phenome” where individual cognitive domains may have distinct genetic and molecular architectures. Further, cognitive tasks vary in their domain specificity and domain overlap [90, 91]. Integrative analytic methods, as discussed below in Section 4.1, may be useful for untangling this complex phenotypic architecture [103]. In addition, low-frequency, rare, and copy number variants may now be readily analyzed using the recently available ADNI WGS data allowing assessment of contributions of these features in cognitive performance.
3.3.8. Multidimensional Data Mining
Besides examining genetic effects of different ADNI phenotypes, some studies have integrated ADNI genetic data with multidimensional phenotypic data sets to jointly predict outcomes of interest. For example, Trzepacz et al. [104] compared the power of three neuroimaging modalities (sMRI, [11C]PiB PET, and [18F]FDG PET), coupled with APOE genotype, for predicting MCI conversion to AD. Zhang et al. [105] compared several feature selection and machine learning algorithms, where ADNI multi-dimensional data sets, including genetic data along with sMRI, [18F]FDG PET, and CSF biomarkers, were used to classify CN, MCI, and AD participants. As mentioned above, multimodal ADNI data sets including APOE ε4 status, CSF biomarkers, MRI, and [18F]FDG PET have been used as predictors to classify AD status [65], predict cognitive decline [66], or predict MCI to AD conversion [67].
4. Discussion
Overall, the many reports evaluating ADNI genetic data using multidimensional phenotypes from rich ADNI data sets confirmed key findings in the genetics of AD and also identified novel candidate genes that deserve further investigation and replication in independent cohorts. Below we discuss several key themes related to the significance and implications of genetic studies using ADNI data as well as plans for the ADNI Genetics Core going forward.
4.1. Toward a systems biology of AD: integrating multi-omics data at the pathway and network level
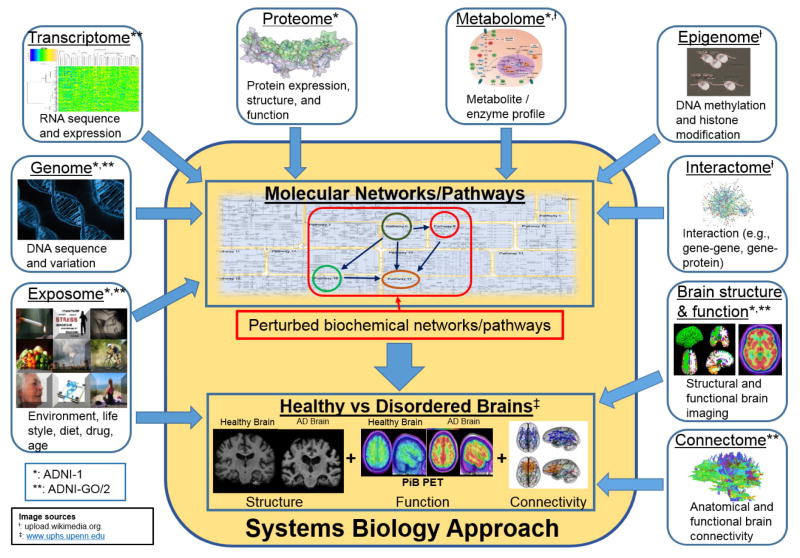
LOAD is thought to have a complex etiology, with multiple genetic and environmental factors presumed to influence susceptibility [106]. This complex model of susceptibility can pose conceptual challenges to the “one-gene/one-enzyme/one-function” paradigm that has been used to characterize disorders with more simple genetic architectures [107, 108]. In response to such challenges, systems biology approaches have been developed that attempt to model complex and interactive multi-level biological systems using a range of advanced computational and experimental tools. These approaches are based on the hypothesis that complex genetic architectures can only be appropriately characterized through analytical strategies designed to capture this complexity [109–111]. In practice, these approaches rely on two fundamental premises, namely that (1) genes and their variants are not isolated inciting entities, but rather exist within broader functional pathways and networks [89], and that (2) diseases and their endophenotypes are not isolated endpoints, but rather are collections of perturbations at multiple biological levels including the genome, epigenome, transcriptome, proteome, and metabolome [103, 112]. Figure 3 illustrates the landscape of multiple “-omics” domains relevant to AD. It is striking that ADNI has collected data spanning a broad range of these domains (indicated by asterisks in the figure).
Figure 3.
Converging “multi-omics” in ADNI
This figure illustrates the landscape of multiple “-omics” domains relevant to AD. Note that ADNI has collected data spanning a broad range of these domains (indicated by asterisks; * = data from ADNI-1, ** = data from ADNI-GO/2). Image sources include upload.wikimedia.org (indicated by †) and www.uphs.upenn.edu (indicated by ≠).
One systems biology approach that has been applied to AD starts with an initial discovery of a gene in one “-omics” realm and progresses to test the effect of this gene in complementary biological systems. This strategy can be particularly effective in characterizing the downstream consequences of a genetic variant to fill in knowledge gaps native to most genotype-phenotype associations. There are emerging proof-of-concept examples of this approach that have utilized ADNI data and it seems likely that this approach could be productively extended to other AD-related genes. One integrative functional genomics report followed up on the immune system gene CD33, which was previously implicated in AD through GWAS [113, 114], by demonstrating SNP-associated changes in CD33 expression (protein), phagocytic performance of CD33-expressing monocytes (cellular function), and amyloid accumulation measured by PET imaging (brain pathology) [55]. It is notable that CD33 is expressed selectively in microglia in AD brain tissue [115], is a modulator of microglial activation, and also constitutes a druggable target that has been investigated in acute myeloid leukemia and very recently in AD [116]. Another example, discussed in Section 3.3.7, used GWAS to discover a novel association of the pro-apoptotic gene FASTKD2 with better episodic memory performance and subsequently related the memory-associated SNP to lower FASTKD2 mRNA expression (transcript), lower CSF levels of fas-mediated apoptotic factors (protein), and higher hippocampal volume and grey matter density (brain structure) [80]. Like CD33, FASTKD2 also shows promise as a druggable target based on earlier work in oncology [81]. Future extensions of this systems biology strategy might additionally leverage the ADNI WGS data, as AD-associated genes are likely to contain rare variants with functional implications in addition to the common variants detected by GWAS [117].
Alternatively, systems biology approaches can start with larger gene sets (pathways or networks) identified through “-omics” analysis and subsequently use the known biological relationships within those groups to converge on one or more gene targets. Two recent examples of this approach, including one utilizing ADNI data [118], identified novel regulatory gene targets through network analysis of post-mortem brain tissue transcriptome data. In one report, a network enriched with immune system and microglia-related genes and upregulated in post-mortem AD brain tissue was found to contain TYROBP (TYRO tyrosine kinase binding protein) as a common regulator of many of the genes in the network [119]. Another report identified networks with perturbed gene expression in AD and APOE ε4 carriers and discovered RNF219 (ring finger protein 219) as a key mediator of these transcriptomic changes [118]. Variants in RNF219 were additionally found to be related to increased amyloid deposition by in vivo PET imaging [118]. Overall, the network strategy is particularly attractive for its ability to highlight broad ensembles of genes serving as effectors that drive key disease mechanisms and thus, may hold strong potential for identification of promising therapeutic targets [120].
With rich phenotype and genotype data now available for participants spanning the continuum from normal aging to clinical AD, the next phase of ADNI described below (Section 4.4) will provide an ideal setting for systems biology approaches to elucidate missing pieces in the understanding of AD pathophysiology and to potentially expand the scope of AD diagnostic and therapeutic strategies. From a risk prediction standpoint, networks exhibiting coexistent genetic variation and biological perturbation would represent prime targets in the development of personalized, burden-based genetic susceptibility tests [121]. For therapeutic development, pathways and networks displaying multi-omics relationships with AD would reduce the search space for rational drug design and may highlight “hub” genes for therapeutic cocktail approaches, such as in the polypharmacy strategies successfully employed for AIDS and various cancers [34, 112].
4.2. Implications for clinical trial design and precision medicine
4.2.1. Use of genetics in clinical trial design
The prospect of a personalized or precision medicine for AD, and for its incorporation in therapeutic trial design, is predicated on the ability to use an individual’s genetic profile to refine predisposition to disease, characteristics such as likely rate of progression, and predicted therapeutic and side effect responses to various therapeutic strategies. At present, the utility of genetic data within regards to AD clinical drug development has been largely limited to APOE. Unfortunately, the past decade of clinical drug development programs for AD-modifying therapies has been burdened with failures. Although these failures may be a result of lack of drug efficacy, the inability to measure drug response in homogenous patient populations has contributed an unquestionable source of variability in these programs. APOE’s strong and well-replicated associations with multiple clinical and functional disease endpoints and quantitative traits such as brain volume, age at disease onset, amyloid deposition, and disease risk provide sufficient effect size and predictive value to serve utility in clinical development programs [5, 56, 122, 123]. APOE is commonly used in clinical trial analyses as a stratifying factor or covariate to adjust for heterogeneity, although independently it has failed to show consistent therapeutic effect on drug response within this context [124, 125]. However, APOE did predict dose related side effects (vasogenic edema and stroke). APOE is also being used to select patients for two large drug trials targeting at-risk but cognitively normal individuals, the Alzheimer’s Prevention Initiative (API) APOE and TOMMORROW trials, where this genotype provides an inexpensive and minimally invasive biomarker to identify individuals with high risk of AD. Both APOE and the adjacent TOMM40 gene are being used for patient selection in the TOMMORROW trial. The small effect sizes of the other more than 20 confirmed markers from large GWAS studies has to date limited their utility for trial design and stratification. Polygenic scoring and related risk aggregation strategies may prove useful in addressing the cumulative contributions of multiple genes and pathways [126, 127]. Similarly, analysis of gene-gene and gene-environment interactions, while increasing complexity, is likely to provide important insights relevant for trial design and analysis. As additional information becomes available from multiple sources, the combinatorial roles of genes in targeted pathways are expected to become increasing important for trial design.
4.2.2. Use of genetics in therapeutic target identification
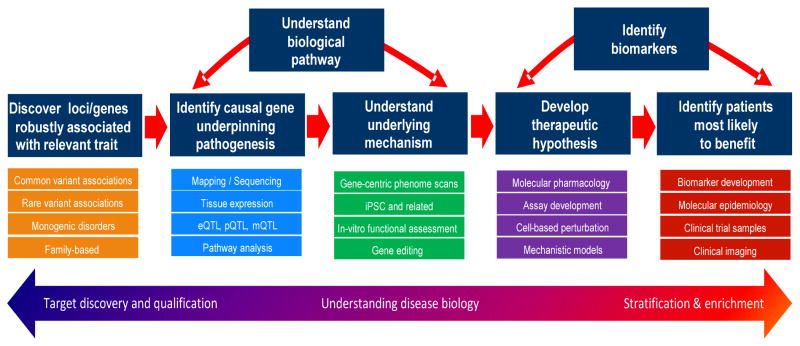
Sources of human variability (genotypic and phenotypic) continue to be characterized in extraordinary detail, scale, and speed, presenting unprecedented opportunities for discovery and development of novel and targeted therapeutic hypotheses. This requires robust insights into relevant human biology. Figure 4 provides an overview of the path from genetic signal detection to targeted therapeutics, including methods and tolls, as well as implications for trial design, as discussed above. The value of genetic and related data in medicine development has been exemplified by a new paradigm for novel and targeted drug discovery in oncology [128, 129]. Three critical tipping points appear to have now been reached for the key principles of this paradigm to be extended to drug discovery and development in neurodegeneration: (1) unprecedented technologies to characterize genotypic and phenotypic causes and consequences of human disease variability are; (2) dense genotype-phenotype data being generated by large-scale public and private sector investments; and, (3) translational systems available to derive novel insights into disease biology and pharmacology from genetic signals. Considerable effort is being invested to leverage and translate human genetic and related data into opportunities for target discovery and validation, understanding disease biology, and patient stratification and enrichment.
Figure 4.
Path from genetic signals to targeted therapeutics: key applications to drug discovery and development
This figure shows an overview of the path from genetic signal detection to targeted therapeutics and implications for trial design. (eQTL = expression quantitative trait loci; pQTL = proteomic quantitative trait loci; mQTL = metabolic quantitative trait loci; iPSC = induced pluripotent stem cells)
Retrospective analyses of drug discovery pipelines indicate that novel therapeutic hypotheses supported by robust human genetic data have a greater likelihood of successfully delivering new, impactful medicines [130]. Strategically, premising drug discovery pipelines upon a foundation of robust human data provides an opportunity for smaller, focused research portfolios of higher confidence targets (contrasting with historical “shots on goal” strategies of pursuing a large number of lower confidence targets with a high rate of attrition [131]). Moreover, advances in approaches to integrate genetic, intermediate trait, and clinical outcome data provide opportunities to distinguish causal relationships from correlative associations based upon the principles of Mendelian Randomization [132]. Within the drug discovery industry, two applications of such integrated data are to identify opportunities for target discovery (“forward genetics”: where a new therapeutic hypothesis is developed subsequent to identification of robust genetic signal) and target qualification (“reverse genetics”: where human genetic data are used to “validate” a therapeutic hypothesis based on non-genetic data, such as pre-clinical animal models).
The growing list of genes identified as being robustly associated with AD has recently identified several potential novel therapeutic targets and target pathways. ADNI data has contributed significantly to such findings. For example, recent genetic association findings highlight autoimmune and inflammation related mechanisms as being potentially causal in AD [133]. Autoimmune mechanisms may provide opportunities for novel therapeutic targets for the treatment or prevention of AD, especially as such mechanisms may provide additional therapeutic value beyond established target pathways such as Aβ and tau [134]. Specific examples of immune-related genes associated with AD risk and/or related intermediate traits include TREM2 and CD33 [55, 135]. The dense genetic, multi-modal phenotypic, and clinical data (including in a longitudinal setting) in ADNI have contributed to better understanding of associations of these genes with AD, including potential pathways and mechanisms involved [55]. As the number of genes and pathways identified as being associated with neurodegenerative diseases grows [34], ADNI provides an important resource to better understand such associations and help contribute to the translation of such associations into therapeutic hypotheses. This contribution may be enhanced given the potential to incorporate functional genomic and mechanistic data in the future.
4.2.3. Use of genetics in novel biomarker identification
Genetic studies are instrumental in elucidating the pathways involved in the etiology of AD and in discovering and validating drug targets. A parallel approach can also be used to identify disease biomarkers. Identification of the TREM2 rare variant and its association with AD risk has sparked renewed interest in the field in understanding the role of inflammation [135]. As this mutation is relatively rare, it likely has little clinical utility as a biomarker of disease heterogeneity. However, in part because of this and other immune related genetic findings, there is renewed appreciation of the need for finding blood biomarkers or imaging methods that track with neuroinflammation. Pathway analysis approaches can be used to identify such biomarker targets, as was demonstrated by Zhang et al [119]. As our understanding of disease complexity in AD matures, genomic science can help to elucidate underlying biology, bringing together multiple levels of “-omics” data to the level of individual variability to uncover novel signals and biomarkers that may segregate with disease subtypes.
4.3. Understanding AD biology through induced pluripotent stem cells (iPSCs)
Cancer research has benefited from renewable sources of cell lines derived from tumors carrying alterations of major oncogenic pathways to enable drug screening and characterization. By contrast, drug discovery in neurodegenerative diseases such as AD have mostly relied on animal models. Advances in iPSC technology have opened new opportunities for creation of novel human-derived in vitro models to help bridge the gap between animal models and clinical development.
iPSC cells can be generated from various adult cell types such as skin fibroblasts, hair follicle keratinocyte, or liver or blood cells. Similar to embryonic stem cells, iPSCs can self-renew and be differentiated into all three germ layers to allow further investigations in relevant tissue/cell types [136]. The first iPSCs for AD were generated from fibroblasts of familial AD patients carrying mutations in PSEN1 or PSEN2 [137]. Neurons differentiated from these iPSCs have elevated Aβ1-42 secretion, which can be modulated by gamma-secretase inhibitors. A later study by Israel et al. [138] created iPSCs using fibroblasts from familial AD, sporadic AD, and non-demented controls. Similar to the earlier report, differentiated neurons from iPSCs from familial and sporadic AD showed higher levels of Aβ and phosphorylated tau compared to those from controls. These studies indicated that it is possible to generate iPSC-derived neurons from elderly patients that capture basic biochemical changes associated with disease states yielding potential in vitro systems for compound screening. Promising recent technical developments also suggest the feasibility of producing iPSCs with appropriate disease phenotypes from LCLs [139]. This could be very important in maximizing the value of ADNI data as LCLs are available for almost every participant from all study phases (ADNI-1/GO/2).
The vast majority of AD cases are sporadic with contributions from multiple genetic and environmental factors. Even though individuals with AD share similar pathological changes, the patient population is highly heterogeneous. The preliminary comparison reported by Israel et al. showed that only one of the two sporadic AD iPSC differentiated neurons shared similar phenotypes with those from familial AD participants, indicating that sporadic AD patients may have different degrees of similarity to familial AD phenotypes. A larger panel of iPSC lines derived from sporadic AD patients may provide further information related to disease heterogeneity and help define potential disease subtypes within sporadic AD. Recent work from the University of California, San Diego group suggests the potential of iPSC studies to identify molecular phenotypic signatures associated with established AD candidate genes such as SORL1 (sortilin-related receptor, L(DLR class) A repeats containing) [140].
ADNI has stimulated international efforts to characterize AD phenotypes across a number of technology platforms including imaging, biochemistry, and genetics. iPSC lines derived from these cohorts, including prodromal dementia participants and cognitively normal at-risk participants with rich characterizations, will enable linking in vitro molecular profiles (e.g., epigenetic changes) with other relevant disease phenotypes. In fact, the next phase of ADNI would be well-positioned to collect samples for iPSC development on a large series of older adults at risk for MCI and AD with highly detailed phenotypic characterization that could help to determine the relationship between clinical and pathophysiological phenotypes and cellular/molecular phenotypes. Recent developments in 3-dimensional (3D) culture systems may permit capture of additional disease features such as neurofibrillary tangles [141]. This will provide better in vitro model systems to understand disease mechanisms that, combined with improved phenotypic characterization of disease heterogeneity, can be expected to advance AD therapeutics and move the field closer to the goal of precision medicine.
4.4. Return of Research Results to Participants
The topic of return of research results (RORR) to research participants and/or their physicians has been a source of considerable interest and controversy across the entire spectrum of the research enterprise [142–144], but particularly in the domains of imaging [145–147] and genetics [148–150]. In brief, as human studies systematically collect information on participants, participants may request the information and arguments may be made for participant benefit of receiving this information. Discussions of RORR sometimes differentiate information that is relevant to the disease in question and the aims of the study (sometimes called “incidental research findings”) versus information unrelated to the purpose of the study (sometimes referred to as “incidental findings”). The discussion also sometimes differentiates RORR that are aligned with clinical standards of care (for example, returning an unexpectedly elevated blood pressure or the presence of an unexpected mass on an imaging study) versus returning results that are not yet part of expected standards of clinical care where the implications of the results may not yet be well-understood (such as new assays or novel imaging modalities).
There have been active discussions about RORR within various ADNI committees and within the ADNI Genetics Core. Institutional review boards at various ADNI clinical sites have also raised questions with regard to how both imaging and genetic RORR issues should be handled. In ADNI, the issue has arisen in two principal areas: amyloid PET imaging and genomics.
4.4.1. RORR for Amyloid PET Imaging
A large subset of ADNI participants received amyloid PET scans as part of the ADNI research protocol. In some of these instances, either the participants themselves have requested their amyloid PET results, or their clinicians (who are sometimes also the ADNI investigators at that particular site) have requested the results of the amyloid PET scans in order to influence the clinical care of these participants – particularly among cognitively normal participants who are found to have positive amyloid PET scans. Arguments against disclosing these data were that they were ordered outside of the clinical work stream, often in situations (such as in unaffected individuals) that were outside the scope of current medical practice, and that they were not all necessarily read in a standardized way by interpreters who had been trained to render a clinical report. A further argument against disclosing these results was that a major component of the scientific value of ADNI is in the correlative analyses between cognitive testing, biomarker collection, structural and functional imaging, and longitudinal outcome. Indeed, it has been demonstrated that simply learning that one has a neurocognitive diagnosis [151], or that one is at increased genetic risk for AD dementia [152], can have a deleterious effect neurocognitive performance. If the results of amyloid PET scans performed under a research protocol were prematurely fed back into the clinical work stream, the choices of clinicians and the reactions of participants could be altered in ways that could diminish the scientific validity of the results.
Arguments for RORR to participants and their clinicians are predominantly focused upon the autonomy of participants and the rights of participants to learn information about themselves that could influence their health [142, 150]. There is also the universal appetite among research participants of all types for the return of health-related research information [153]. In just the arena of dementia, a nationally representative survey of United States adults [154], and an entirely separate survey of adults in the United States and four European countries [155], showed that nearly 70% of those queried would take a test to learn more about their risk of AD, even if no preventative treatment was available. In a 2012 survey of 260 academic physicians specializing in dementia care and research, 135 (52%) responded, and of those, 24% reported that if amyloid scans were FDA approved and covered by insurance, they planned to use this technology for screening asymptomatic individuals [156]. Since that time, published guidelines have recommended against amyloid scans in cognitively normal individuals [157, 158] and United States Centers for Medicare and Medicaid Services (CMS) issued a decision denying payment for clinical use of this technology [159, 160]. Of note, CMS approved coverage with evidence development (CED) for amyloid PET in which patients meeting appropriate use criteria can receive coverage if they are enrolled in an approved comparative effectiveness trial designed to clarify the benefit to clinical care. In our 2012 survey of ADNI investigators, asking respondents to assume FDA approval of [18F]Florbetapir, over half of the 159 respondents indicated that cognitively normal study participants had requested the results of their amyloid scans and 9 respondents (6%) reported that they had already disclosed amyloid imaging results to these participants [161]. Within ADNI, there was a considerable range of opinion regarding the policy that should be adopted for RORR of amyloid PET scans, and in 2012 a policy was adopted that recommended against the RORR for this modality (http://www.adni-info.org/scientists/doc/10_Green_RORR.pdf). These issues are likely to be revisited periodically in this rapidly evolving area.
4.4.2. RORR for Biomarkers and Genetic Variants
In addition to imaging studies, ADNI generates individual level data on a variety of serum, plasma and cerebrospinal fluid biomarkers (see ADNI Biomarker Core report in this Special Issue), as well as the genotype and sequencing results described earlier in this paper. In the areas of biomarkers and genetic variants related to AD, the arguments for and against disclosure are similar to those made about research imaging results, i.e. respect for participant autonomy and possible benefit to ongoing and future medical care versus the possibility that longitudinal measures of important outcomes could be biased by RORR or by actions taken in response to RORR. In the area of laboratory tests there is another consideration, which is that most of the biomarker and genetic data is generated in laboratories that are not CLIA-certified. CLIA is the Clinical Laboratory Improvement Amendments certification by CMS that regulates quality assurance in human clinical laboratory testing in the United States. The position of CMS, and of most Institutional Review Boards (IRBs), is quite unambiguously that no health-related laboratory results should be returned to participants unless those results are generated (or replicated) in a CLIA-certified laboratory [162, 163].
As large scale studies like ADNI collect and analyze genomic information, the societal trends toward return of research results and individual empowerment are encouraging extensive discussion about whether specific processes should be put in place to return incidental or secondary genomic findings of medical importance [150, 164]. Similar discussions have been occurring in the clinical realm as whole exome and whole genome sequencing are increasingly being utilized in clinical diagnosis [165, 166], sparked in part by the recommendations of the American College of Medical Genetics and Genomics (ACMG) that were published in 2013 and updated in 2015 [167, 168]. The ACMG recommended that an explicit panel of mutations in specific genes should be routinely assessed when sequencing is performed for clinical purposes. The ACMG recommendations recognized that this area is evolving and suggested a need for regular re-evaluations of the minimum list, as well as the importance of clear communication with the patient, but attempted to place the return of secondary findings in clinical sequencing squarely within existing traditions of clinical medicine [169]. These discussions in the clinical arena have prompted extensive considerations by researchers and IRBs about the distinctions between the clinical and research domains [162, 163], but consensus recommendations appear to be evolving toward the position that genomic results of medical importance and medical actionability should be shared with research participants and after the death of participants, with their families, particularly if the consent forms are clearly written to include this contingency [150, 170].
Large-scale genotyping and sequencing in a dementia study raises a number of other issues. Many individuals and families are interested in learning their APOE genotype, as this is a risk marker for AD that has become widely recognized throughout society. APOE genotype is a robust marker for AD risk, but does not meet the criteria for medical importance and medical actionability that have been described in the above referenced discussions of RORR. Thus, while clinical trials have demonstrated that APOE genotype can be returned without undue distress [171–174], this is not typically included in discussions of possible genomic RORR.
Both imaging and genome-scale testing also raise issues of re-identifiability. With high resolution MRI scanning, it is theoretically possible to reconstruct faces to the point where they might be identifiable. There have also been well-publicized accounts in which de-identified genomes could be linked to specific surnames, and potentially to specific individuals, through linkage to publically available databases [175]. In this regard, ADNI genomes are as vulnerable, or protected, as other genomes that are being made available to qualified researchers through various sharing mechanisms such as dbGaP. While re-identification through imaging or genomic information is therefore possible, it is strictly prohibited by ADNI policies and by IRBs, and any attempt to do so by investigators would be in clear violation of these policies.
In summary, the RORR to participants in ADNI has not been pursued to date because ADNI consent forms do not promise that incidental findings will be returned and in some cases are explicit in noting that they will not be returned. It remains to be seen if IRB policies toward return of incidental genomic information will request or require ADNI sites to revisit this question in the years ahead.
4.5. Future plans, related initiatives, and conclusions
ADNI-2 is now about to enter its final year and plans are being finalized for the ADNI-3 proposal to continue and extend this research in important new directions. Tau PET imaging studies, computer-based cognitive assessments, and other new features will be added this year and hopefully carried forward into the next phase of ADNI. From a genetic perspective, these novel phenotypes will be important topics for future association studies. In addition to continued banking, dissemination, and assays on longitudinal genetic biosample data, major plans include analysis of existing baseline and follow-up visit data and similar collection and analysis of new participants to be recruited in the future. Most resources available to the Genetics Core are for sample collection and processing. However, analysis of existing data on the association of genetic variation and key clinical/cognitive, imaging, and biomarker phenotypes is a priority. Longitudinal blood RNA and epigenetic markings (including but not limited to methylation) in DNA will be important and potentially highly informative. As discussed above, a longitudinal convergent “multi-omics” collection and analysis strategy will continue, along with the challenge of finding resources for complete analysis and dissemination of all biosample results. Partnerships with the Alzheimer’s Association, other foundations and donors, and ADNI’s industry partners via the Foundation for the National Institutes of Health have made possible the early ADNI-1 GWAS, the recent WGS on over 800 samples, and the RNA expression profiling using blood samples from most of the same participants.
The ADNI Genetics Core hosts an extended Working Group that holds conference calls monthly or more often as needed. The Genetics Core also sponsors an ad hoc RNA Working Group to support collaborative expression profiling and transcriptome analysis. Further, the Genetics Core has recently established an ADNI Systems Biology Working Group to enhance the potential contributions of this specialized area for analysis of ADNI data and for collaboration on analyses that span multiple data types and cohorts. Thought leaders in systems biology and network medicine from academia, foundations, and industry are engaged in determining the key scientific questions, opportunities, and priorities that these areas of science can contribute to ADNI.
The ADNI Genetics Core has served as a liaison to the relevant consortia including the Alzheimer’s Disease Genetics Consortium (ADGC)/Alzheimer’s Disease Sequencing Project (ADSP), ROS/MAP, AddNeuroMed, World-Wide ADNI (WW-ADNI), Enhancing Neuro Imaging Genetics through Meta-Analysis (ENIGMA), Multi-Institutional Research in Alzheimer’s Genetic Epidemiology (MIRAGE), IMAS, and HRS investigators, as well as other cohort studies that could serve as replication samples or collaborate in other synergistic research activities. Various inquiries about ADNI genetic data within and outside of ADNI have been addressed to encourage the individual and collaborative investigation of ADNI genetic data. Current plans include direct comparisons between ADNI and the autosomal dominant form of AD seen in the Dominantly Inherited Alzheimer Network (DIAN) Consortium. One limitation of ADNI genetic analyses is the absence of large data sets for replication with the vast array of imaging and fluid biomarkers available in ADNI. To date, some studies have utilized local data sets not publically shared, as well as some partially publically available data sets. Additional open data sharing in the future will assist in providing replication samples for ADNI genetic analyses. Further, when genetic data become available for the Australian Imaging, Biomarker & Lifestyle Flagship Study of Ageing (AIBL) Study and other publically available cohorts in the WW-ADNI consortium there will be tremendously enhanced power to detect associations and replicate ADNI genetic findings.
As promising leads are discovered using ADNI genetic and phenotypic data, there is potentially very high scientific and translational value to be gained from follow-up with functional genomics experiments (as illustrated in Figure 4). One particularly promising direction for functional genomics follow-on studies is creation of iPSCs from blood or fibroblast samples collected from the ADNI cohort. iPSCs coupled with other clinical and imaging information from the same individual would help to construct “dementia-on-a-chip” models for assessing candidate diagnostic and therapeutic agents. Although it is beyond the scope of ADNI to follow-up all promising leads, the Genetics Core will continue to facilitate and coordinate with partners and the research community to enable these experiments to be conducted and results shared expeditiously. Finally, given the complexity of rich longitudinal “multi-omics” phenotypes, intelligent analytic strategies closely linked to biological knowledge need to be adapted and applied to ADNI data. Fortunately, the computational science community is stepping up to this challenge with dozens of publications employing advanced strategies to interrogate ADNI data and the Genetics Core looks forward to supporting and collaborating with such efforts to extract biologically significant and clinically relevant information from the proverbial haystack of information.
Supplementary Material
Acknowledgments
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; ; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. Samples from the National Cell Repository for Alzheimer’s Disease (NCRAD), which receives government support under a cooperative agreement (U24AG021886) awarded by the National Institute on Aging, were used in this study. Data analysis was supported in part by NIH R01 LM011360, U01 AG024904, RC2 AG036535, R01 AG19771, P30 AG10133, UL1TR001108, DOD W81XWH-14-2-0151, NCAA 14132004, the Alzheimer’s Association, and NSF IIS-1117335.
Abbreviations
- 3D
3-dimensional
- Aβ
amyloid-beta
- Aβ1-42
amyloid-beta 1-42
- ACE
angiotensin-converting enzyme
- ACMG
American College of Medical Genetics and Genomics
- AD
Alzheimer’s Disease
- ADGC
Alzheimer’s Disease Genetics Consortium
- ADNI
the Alzheimer’s Disease Neuroimaging Initiative
- ADSP
Alzheimer’s Disease Sequencing Project (ADSP)
- AIBL
Australian Imaging, Biomarker & Lifestyle Flagship Study of Ageing
- API
Alzheimer’s Prevention Initiative
- APOE
apolipoprotein E
- ARRA
American Recovery and Reinvestment Act of 2009
- ATP
adenosine triphosphate
- BLSA
Baltimore Longitudinal Study of Aging
- BMS
Bristol-Myers Squibb
- CC
C-C motif
- CFHR1
complement factor H-related protein 1
- CLIA
Clinical Laboratory Improvement Amendments
- CMS
Centers for Medicare and Medicaid Services
- CNV
copy number variant
- CSER
Clinical Sequencing Exploratory Research Consortium
- CSF
cerebrospinal fluid
- DIAN
Dominantly Inherited Alzheimer Network
- DNA
deoxyribonucleic acid
- DTI
diffusion tensor imaging
- EDTA
ethylenediaminetetraacetic acid
- ELSI
ethical, legal, and social implications
- EMCI
early mild cognitive impairment
- eMERGE
Electronic Medical Records and Genomics Network
- ENIGMA
Enhancing Neuro Imaging Genetics through Meta-Analysis
- FDA
Food and Drug Administration
- FDG
[18F]Fluorodeoxyglucose
- fMRI
functional magnetic resonance imaging
- GWAS
genome-wide association study
- HCI
hypometabolic convergence index
- HRS
United States Health and Retirement Study
- ILR6
interleukin-6 receptor (IL6R)
- IMAS
Indiana Memory and Aging Study
- iPSC
induced pluripotent stem cells
- IRB
Institutional Review Board
- LCL
lymphoblastoid cell lines
- LMCI
late mild cognitive impairment
- LOAD
late-onset Alzheimer’s disease
- LONI
Laboratory of Neuro Imaging
- MAP
Rush Memory and Aging Project
- MCI
mild cognitive impairment
- MIRAGE
Multi-Institutional Research in Alzheimer’s Genetic Epidemiology
- MMP-3
matrix metalloproteinase-3
- MRI
magnetic resonance imaging
- NCRAD
National Cell Repository for Alzheimer’s Disease
- NGS
next generation sequencing
- NIA
National Institute on Aging
- NIBIB
National Institute of Biomedical Imaging and Bioengineering
- NIH
National Institutes of Health
- PCR
polymerase chain reaction
- PET
positron emission tomography
- PiB
[11C]Pittsburgh Compound B
- pTau181
Tau phosphorylated at threonine (position 181)
- QC
quality control
- QTL
quantitative trait loci
- RNA
ribonucleic acid
- ROR
return of results
- ROS
Religious Order Study
- SCD
subjective cognitive decline
- SMC
significant memory concern
- sMRI
structural magnetic resonance imaging
- SNP
single nucleotide polymorphism
- WW-ADNI
world-wide Alzheimer’s Disease Neuroimaging Initiative
- WBC
white blood cells
- WES
whole exome sequencing
- WGS
whole genome sequencing
Footnotes
5. Supplementary Information: Endnote database of ADNI Genetics Publications (ADNI_Genetics_papers_2008-2014.enlx)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Risacher SL, Kim S, Nho K, Foroud TM, Shen L, Petersen RC, et al. APOE effect on Alzheimer’s biomarkers in older adults with significant memory concern. Alzheimer’s and Dementia. 2015 doi: 10.1016/j.jalz.2015.03.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jessen F, Amariglio RE, van Boxtel M, Breteler M, Ceccaldi M, Chetelat G, et al. A conceptual framework for research on subjective cognitive decline in preclinical Alzheimer’s disease. Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2014;10:844–52. doi: 10.1016/j.jalz.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. The Alzheimer’s Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 2013;9:e111–94. doi: 10.1016/j.jalz.2013.05.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman RJ, Aisen PS, De Strooper B, Fox NC, Lemere CA, Ringman JM, et al. Autosomal-dominant Alzheimer’s disease: a review and proposal for the prevention of Alzheimer’s disease. Alzheimer’s research & therapy. 2011;3:1. doi: 10.1186/alzrt59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nature genetics. 2013;45:1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potkin SG, Turner JA, Guffanti G, Lakatos A, Torri F, Keator DB, et al. Genome-wide strategies for discovering genetic influences on cognition and cognitive disorders: methodological considerations. Cognitive neuropsychiatry. 2009;14:391–418. doi: 10.1080/13546800903059829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–50. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 8.Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S, et al. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2010;6:265–73. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, et al. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer’s disease. PloS one. 2009;4:e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen L, Thompson PM, Potkin SG, Bertram L, Farrer LA, Foroud TM, et al. Genetic analysis of quantitative phenotypes in AD and MCI: imaging, cognition and biomarkers. Brain imaging and behavior. 2014;8:183–207. doi: 10.1007/s11682-013-9262-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reymer PW, Groenemeyer BE, van de Burg R, Kastelein JJ. Apolipoprotein E genotyping on agarose gels. Clinical chemistry. 1995;41:1046–7. [PubMed] [Google Scholar]
- 12.Hixson JE, Vernier DT. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. Journal of lipid research. 1990;31:545–8. [PubMed] [Google Scholar]
- 13.Hawkins JR, Khripin Y, Valdes AM, Weaver TA. Miniaturized sealed-tube allele-specific PCR. Human mutation. 2002;19:543–53. doi: 10.1002/humu.10060. [DOI] [PubMed] [Google Scholar]
- 14.Myakishev MV, Khripin Y, Hu S, Hamer DH. High-throughput SNP genotyping by allele-specific PCR with universal energy-transfer-labeled primers. Genome research. 2001;11:163–9. doi: 10.1101/gr.157901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roses AD. An inherited variable poly-T repeat genotype in TOMM40 in Alzheimer disease. Archives of neurology. 2010;67:536–41. doi: 10.1001/archneurol.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nho K, Corneveaux JJ, Kim S, Lin H, Risacher SL, Shen L, et al. Identification of functional variants from whole-exome sequencing, combined with neuroimaging genetics. Molecular psychiatry. 2013;18:739. doi: 10.1038/mp.2013.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nho K, Corneveaux JJ, Kim S, Lin H, Risacher SL, Shen L, et al. Whole-exome sequencing and imaging genetics identify functional variants for rate of change in hippocampal volume in mild cognitive impairment. Molecular psychiatry. 2013;18:781–7. doi: 10.1038/mp.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irizarry RA, Hobbs B, Collin F, Beazer - Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 19.Vawter MP, Evans S, Choudary P, Tomita H, Meador-Woodruff J, Molnar M, et al. Gender-specific gene expression in post-mortem human brain: localization to sex chromosomes. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2004;29:373–84. doi: 10.1038/sj.npp.1300337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schadt EE, Woo S, Hao K. Bayesian method to predict individual SNP genotypes from gene expression data. Nature genetics. 2012;44:603–8. doi: 10.1038/ng.2248. [DOI] [PubMed] [Google Scholar]
- 21.Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell. 2008;134:714–7. doi: 10.1016/j.cell.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 22.Kaddurah-Daouk R, Krishnan KR. Metabolomics: a global biochemical approach to the study of central nervous system diseases. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2009;34:173–86. doi: 10.1038/npp.2008.174. [DOI] [PubMed] [Google Scholar]
- 23.Kaddurah-Daouk R, Weinshilboum RM. Pharmacometabolomics: implications for clinical pharmacology and systems pharmacology. Clinical pharmacology and therapeutics. 2014;95:154–67. doi: 10.1038/clpt.2013.217. [DOI] [PubMed] [Google Scholar]
- 24.Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nature reviews Molecular cell biology. 2012;13:263–9. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han X, Rozen S, Boyle SH, Hellegers C, Cheng H, Burke JR, et al. Metabolomics in early Alzheimer’s disease: identification of altered plasma sphingolipidome using shotgun lipidomics. PloS one. 2011;6:e21643. doi: 10.1371/journal.pone.0021643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaddurah-Daouk R, Rozen S, Matson W, Han X, Hulette CM, Burke JR, et al. Metabolomic changes in autopsy-confirmed Alzheimer’s disease. Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2011;7:309–17. doi: 10.1016/j.jalz.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Motsinger-Reif AA, Zhu H, Kling MA, Matson W, Sharma S, Fiehn O, et al. Comparing metabolomic and pathologic biomarkers alone and in combination for discriminating Alzheimer’s disease from normal cognitive aging. Acta neuropathologica communications. 2013;1:28. doi: 10.1186/2051-5960-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S, Swaminathan S, Inlow M, Risacher SL, Nho K, Shen L, et al. Influence of genetic variation on plasma protein levels in older adults using a multi-analyte panel. PloS one. 2013;8:e70269. doi: 10.1371/journal.pone.0070269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boada M, Antunez C, Ramirez-Lorca R, DeStefano AL, Gonzalez-Perez A, Gayan J, et al. ATP5H/KCTD2 locus is associated with Alzheimer’s disease risk. Molecular psychiatry. 2014;19:682–7. doi: 10.1038/mp.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leduc V, De Beaumont L, Theroux L, Dea D, Aisen P, Petersen RC, et al. HMGCR is a genetic modifier for risk, age of onset and MCI conversion to Alzheimer’s disease in a three cohorts study. Molecular psychiatry. 2014 doi: 10.1038/mp.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biffi A, Sabuncu MR, Desikan RS, Schmansky N, Salat DH, Rosand J, et al. Genetic variation of oxidative phosphorylation genes in stroke and Alzheimer’s disease. Neurobiology of aging. 2014;35:1956.e1–8. doi: 10.1016/j.neurobiolaging.2014.01.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim JH, Song P, Lim H, Lee JH, Lee JH, Park SA, et al. Gene-based rare allele analysis identified a risk gene of Alzheimer’s disease. PloS one. 2014;9:e107983. doi: 10.1371/journal.pone.0107983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desikan RS, Schork AJ, Wang Y, Witoelar A, Sharma M, McEvoy LK, et al. Genetic overlap between Alzheimer’s disease and Parkinson’s disease at the MAPT locus. Molecular psychiatry. 2015 doi: 10.1038/mp.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramanan VK, Saykin AJ. Pathways to neurodegeneration: mechanistic insights from GWAS in Alzheimer’s disease, Parkinson’s disease, and related disorders. American journal of neurodegenerative disease. 2013;2:145–75. [PMC free article] [PubMed] [Google Scholar]
- 35.Nho K, Kim S, Risacher SL, Shen L, Corneveaux JJ, Swaminathan S, et al. Protective variant for hippocampal atrophy identified by whole exome sequencing. Annals of neurology. 2015;77:547–52. doi: 10.1002/ana.24349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nho K, Saykin AJ. Reply to Letter to the Editor. Annals of neurology. 2015 doi: 10.1002/ana.24480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu T, Aron L, Zullo J, Pan Y, Kim H, Chen Y, et al. REST and stress resistance in ageing and Alzheimer’s disease. Nature. 2014;507:448–54. doi: 10.1038/nature13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hohman TJ, Koran ME, Thornton-Wells TA Alzheimer’s Disease Neuroimaging I. Genetic modification of the relationship between phosphorylated tau and neurodegeneration. Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2014 doi: 10.1016/j.jalz.2013.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hohman TJ, Koran ME, Thornton-Wells TA Alzheimer’s Neuroimaging I. Genetic variation modifies risk for neurodegeneration based on biomarker status. Frontiers in aging neuroscience. 2014;6:183. doi: 10.3389/fnagi.2014.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roussotte FF, Gutman BA, Hibar DP, Madsen SK, Narr KL, Thompson PM, et al. Carriers of a common variant in the dopamine transporter gene have greater dementia risk, cognitive decline, and faster ventricular expansion. Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2014 doi: 10.1016/j.jalz.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roussotte FF, Gutman BA, Madsen SK, Colby JB, Thompson PM Alzheimer’s Disease Neuroimaging I. Combined effects of Alzheimer risk variants in the CLU and ApoE genes on ventricular expansion patterns in the elderly. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2014;34:6537–45. doi: 10.1523/JNEUROSCI.5236-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roussotte FF, Jahanshad N, Hibar DP, Sowell ER, Kohannim O, Barysheva M, et al. A commonly carried genetic variant in the delta opioid receptor gene, OPRD1, is associated with smaller regional brain volumes: replication in elderly and young populations. Human brain mapping. 2014;35:1226–36. doi: 10.1002/hbm.22247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roussotte FF, Gutman BA, Hibar DP, Jahanshad N, Madsen SK, Jack CR, Jr, et al. A single nucleotide polymorphism associated with reduced alcohol intake in the RASGRF2 gene predicts larger cortical volumes but faster longitudinal ventricular expansion in the elderly. Frontiers in aging neuroscience. 2013;5:93. doi: 10.3389/fnagi.2013.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luis EO, Ortega-Cubero S, Lamet I, Razquin C, Cruchaga C, Benitez BA, et al. Frontobasal gray matter loss is associated with the TREM2 p.R47H variant. Neurobiology of aging. 2014 doi: 10.1016/j.neurobiolaging.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajagopalan P, Hibar DP, Thompson PM. TREM2 and neurodegenerative disease. The New England journal of medicine. 2013;369:1565–7. doi: 10.1056/NEJMc1306509#SA3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koran ME, Hohman TJ, Meda SA, Thornton-Wells TA. Genetic interactions within inositol-related pathways are associated with longitudinal changes in ventricle size. Journal of Alzheimer’s disease: JAD. 2014;38:145–54. doi: 10.3233/JAD-130989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andrawis JP, Hwang KS, Green AE, Kotlerman J, Elashoff D, Morra JH, et al. Effects of ApoE4 and maternal history of dementia on hippocampal atrophy. Neurobiology of aging. 2012;33:856–66. doi: 10.1016/j.neurobiolaging.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Apostolova LG, Hwang KS, Kohannim O, Avila D, Elashoff D, Jack CR, Jr, et al. ApoE4 effects on automated diagnostic classifiers for mild cognitive impairment and Alzheimer’s disease. NeuroImage Clinical. 2014;4:461–72. doi: 10.1016/j.nicl.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jahanshad N, Rajagopalan P, Hua X, Hibar DP, Nir TM, Toga AW, et al. Genome-wide scan of healthy human connectome discovers SPON1 gene variant influencing dementia severity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4768–73. doi: 10.1073/pnas.1216206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warstadt NM, Dennis EL, Jahanshad N, Kohannim O, Nir TM, McMahon KL, et al. Serum cholesterol and variant in cholesterol-related gene CETP predict white matter microstructure. Neurobiology of aging. 2014 doi: 10.1016/j.neurobiolaging.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jahanshad N, Bhatt P, Hibar DP, Villalon JE, Nir TM, Toga AW, et al. Bivariate genome-wide association study of genetically correlated neuroimaging phenotypes from DTI and MRI through a Seemingly Unrelated Regression model. MICCAI MBIA Workshop; 2013; Nagoya, Japan. 2013. [Google Scholar]
- 52.Jahanshad N, Kochunov P, Glahn D, Blangero J, Nichols TE, McMahon KL, et al. Power Estimates for Voxel-Based Genetic Association Studies using Diffusion Imaging. MICCAI MBIA Workshop; 2013; Nagoya, Japan. 2013. [Google Scholar]
- 53.Warstadt NM, Jahanshad N, Dennis EL, Kohannim O, McMahon KL, De Zubicaray GI, et al. Identifying Candidate Gene Effects by Restricting Search Space in a Multivariate Genetic Analysis of White Matter Microstructure. IEEE International Symposium on Biomedical Imaging; Beijing, China. 2014. [Google Scholar]
- 54.Swaminathan S, Shen L, Risacher SL, Yoder KK, West JD, Kim S, et al. Amyloid pathway-based candidate gene analysis of [(11)C]PiB-PET in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort. Brain imaging and behavior. 2012;6:1–15. doi: 10.1007/s11682-011-9136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bradshaw EM, Chibnik LB, Keenan BT, Ottoboni L, Raj T, Tang A, et al. CD33 Alzheimer’s disease locus: altered monocyte function and amyloid biology. Nature neuroscience. 2013;16:848–50. doi: 10.1038/nn.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramanan VK, Risacher SL, Nho K, Kim S, Swaminathan S, Shen L, et al. APOE and BCHE as modulators of cerebral amyloid deposition: a florbetapir PET genome-wide association study. Molecular psychiatry. 2014;19:351–7. doi: 10.1038/mp.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thambisetty M, An Y, Nalls M, Sojkova J, Swaminathan S, Zhou Y, et al. Effect of complement CR1 on brain amyloid burden during aging and its modification by APOE genotype. Biological psychiatry. 2013;73:422–8. doi: 10.1016/j.biopsych.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swaminathan S, Risacher SL, Yoder KK, West JD, Shen L, Kim S, et al. Association of plasma and cortical amyloid beta is modulated by APOE epsilon4 status. Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2014;10:e9–e18. doi: 10.1016/j.jalz.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murphy KR, Landau SM, Choudhury KR, Hostage CA, Shpanskaya KS, Sair HI, et al. Mapping the effects of ApoE4, age and cognitive status on 18F-florbetapir PET measured regional cortical patterns of beta-amyloid density and growth. NeuroImage. 2013;78:474–80. doi: 10.1016/j.neuroimage.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Risacher SL, Kim S, Shen L, Nho K, Foroud T, Green RC, et al. The role of apolipoprotein E (APOE) genotype in early mild cognitive impairment (E-MCI) Frontiers in aging neuroscience. 2013;5:11. doi: 10.3389/fnagi.2013.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jack CR, Jr, Knopman DS, Jagust WJ, Petersen RC, Weiner MW, Aisen PS, et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. The Lancet Neurology. 2013;12:207–16. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan J, Du L, Kim S, Risacher SL, Huang H, Moore JH, et al. Transcriptome-guided amyloid imaging genetic analysis via a novel structured sparse learning algorithm. Bioinformatics. 2014;30:i564–i71. doi: 10.1093/bioinformatics/btu465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hohman TJ, Koran ME, Thornton-Wells TA Alzheimer’s Neuroimaging I. Interactions between GSK3beta and amyloid genes explain variance in amyloid burden. Neurobiology of aging. 2014;35:460–5. doi: 10.1016/j.neurobiolaging.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schraml F, Chen K, Ayutyanont N, Auttawut R, Langbaum JB, Lee W, et al. Association between an Alzheimer’s Disease-Related Index and Gene Dose. PloS one. 2013;8:e67163. doi: 10.1371/journal.pone.0067163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gray KR, Aljabar P, Heckemann RA, Hammers A, Rueckert D Alzheimer’s Disease Neuroimaging I. Random forest-based similarity measures for multi-modal classification of Alzheimer’s disease. NeuroImage. 2013;65:167–75. doi: 10.1016/j.neuroimage.2012.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shaffer JL, Petrella JR, Sheldon FC, Choudhury KR, Calhoun VD, Edward Coleman R, et al. Predicting cognitive decline in subjects at risk for Alzheimer disease by using combined cerebrospinal fluid, MR imaging, and PET biomarkers. Radiology. 2013;266:583–91. doi: 10.1148/radiol.12120010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gomar JJ, Conejero-Goldberg C, Davies P, Goldberg TE Alzheimer’s Disease Neuroimaging I. Extension and refinement of the predictive value of different classes of markers in ADNI: Four-year follow-up data. Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2014 doi: 10.1016/j.jalz.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cruchaga C, Kauwe JS, Harari O, Jin SC, Cai Y, Karch CM, et al. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer’s disease. Neuron. 2013;78:256–68. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chouraki V, De Bruijn RF, Chapuis J, Bis JC, Reitz C, Schraen S, et al. A genome-wide association meta-analysis of plasma Abeta peptides concentrations in the elderly. Molecular psychiatry. 2014;19:1326–35. doi: 10.1038/mp.2013.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kauwe JS, Bailey MH, Ridge PG, Perry R, Wadsworth ME, Hoyt KL, et al. Genome-Wide Association Study of CSF Levels of 59 Alzheimer’s Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation. PLoS genetics. 2014;10:e1004758. doi: 10.1371/journal.pgen.1004758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramirez A, van der Flier WM, Herold C, Ramonet D, Heilmann S, Lewczuk P, et al. SUCLG2 identified as both a determinator of CSF Abeta1-42 levels and an attenuator of cognitive decline in Alzheimer’s disease. Human molecular genetics. 2014 doi: 10.1093/hmg/ddu372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Toledo JB, Bjerke M, Da X, Landau SM, Foster NL, Jagust W, et al. Nonlinear Association Between Cerebrospinal Fluid and Florbetapir F-18 beta-Amyloid Measures Across the Spectrum of Alzheimer Disease. JAMA neurology. 2015 doi: 10.1001/jamaneurol.2014.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang YL, Fennema-Notestine C, Holland D, McEvoy LK, Stricker NH, Salmon DP, et al. APOE interacts with age to modify rate of decline in cognitive and brain changes in Alzheimer’s disease. Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2014;10:336–48. doi: 10.1016/j.jalz.2013.05.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.De Jager PL, Shulman JM, Chibnik LB, Keenan BT, Raj T, Wilson RS, et al. A genome-wide scan for common variants affecting the rate of age-related cognitive decline. Neurobiology of aging. 2012;33:1017.e1–15. doi: 10.1016/j.neurobiolaging.2011.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Donohue MC, Sperling RA, Salmon DP, Rentz DM, Raman R, Thomas RG, et al. The preclinical Alzheimer cognitive composite: measuring amyloid-related decline. JAMA neurology. 2014;71:961–70. doi: 10.1001/jamaneurol.2014.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mormino EC, Betensky RA, Hedden T, Schultz AP, Ward A, Huijbers W, et al. Amyloid and APOE epsilon4 interact to influence short-term decline in preclinical Alzheimer disease. Neurology. 2014;82:1760–7. doi: 10.1212/WNL.0000000000000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kantarci K, Lowe V, Przybelski SA, Weigand SD, Senjem ML, Ivnik RJ, et al. APOE modifies the association between Abeta load and cognition in cognitively normal older adults. Neurology. 2012;78:232–40. doi: 10.1212/WNL.0b013e31824365ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barral S, Bird T, Goate A, Farlow MR, Diaz-Arrastia R, Bennett DA, et al. Genotype patterns at PICALM, CR1, BIN1, CLU, and APOE genes are associated with episodic memory. Neurology. 2012;78:1464–71. doi: 10.1212/WNL.0b013e3182553c48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sherva R, Tripodis Y, Bennett DA, Chibnik LB, Crane PK, de Jager PL, et al. Genome-wide association study of the rate of cognitive decline in Alzheimer’s disease. Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2014;10:45–52. doi: 10.1016/j.jalz.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ramanan VK, Nho K, Shen L, Risacher SL, Kim S, McDonald BC, et al. FASTKD2 is associated with memory and hippocampal structure in older adults. Molecular psychiatry. 2014 doi: 10.1038/mp.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ramanan VK, Saykin AJ. FASTKD2 and human memory: functional pathways and prospects for novel therapeutic target development for Alzheimer’s disease and age-associated memory decline. Pharmacogenomics. 2015;16:429–32. doi: 10.2217/pgs.15.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivieres S, Jahanshad N, et al. Common genetic variants influence human subcortical brain structures. Nature. 2015;520:224–9. doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bakken TE, Roddey JC, Djurovic S, Akshoomoff N, Amaral DG, Bloss CS, et al. Association of common genetic variants in GPCPD1 with scaling of visual cortical surface area in humans. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3985–90. doi: 10.1073/pnas.1105829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stein JL, Hua X, Lee S, Ho AJ, Leow AD, Toga AW, et al. Voxelwise genome-wide association study (vGWAS) NeuroImage. 2010;53:1160–74. doi: 10.1016/j.neuroimage.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Benitez BA, Karch CM, Cai Y, Jin SC, Cooper B, Carrell D, et al. The PSEN1, p.E318G variant increases the risk of Alzheimer’s disease in APOE-epsilon4 carriers. PLoS genetics. 2013;9:e1003685. doi: 10.1371/journal.pgen.1003685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramanan VK, Kim S, Holohan K, Shen L, Nho K, Risacher SL, et al. Genome-wide pathway analysis of memory impairment in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort implicates gene candidates, canonical pathways, and networks. Brain imaging and behavior. 2012;6:634–48. doi: 10.1007/s11682-012-9196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mukherjee S, Kim S, Ramanan VK, Gibbons LE, Nho K, Glymour MM, et al. Gene-based GWAS and biological pathway analysis of the resilience of executive functioning. Brain imaging and behavior. 2014;8:110–8. doi: 10.1007/s11682-013-9259-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nho K, Ramanan VK, Horgusluoglu E, Kim S, Inlow MH, Risacher SL, et al. Comprehensive Gene- and Pathway-Based Analysis of Depressive Symptoms in Older Adults. Journal of Alzheimer’s disease: JAD. 2015 doi: 10.3233/JAD-148009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ramanan VK, Shen L, Moore JH, Saykin AJ. Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends in genetics: TIG. 2012;28:323–32. doi: 10.1016/j.tig.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Harris SE, Deary IJ. The genetics of cognitive ability and cognitive ageing in healthy older people. Trends in cognitive sciences. 2011;15:388–94. doi: 10.1016/j.tics.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 91.Papassotiropoulos A, de Quervain DJ. Genetics of human episodic memory: dealing with complexity. Trends in cognitive sciences. 2011;15:381–7. doi: 10.1016/j.tics.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 92.Rietveld CA, Medland SE, Derringer J, Yang J, Esko T, Martin NW, et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340:1467–71. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Caselli RJ, Dueck AC, Osborne D, Sabbagh MN, Connor DJ, Ahern GL, et al. Longitudinal modeling of age-related memory decline and the APOE epsilon4 effect. The New England journal of medicine. 2009;361:255–63. doi: 10.1056/NEJMoa0809437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schiepers OJ, Harris SE, Gow AJ, Pattie A, Brett CE, Starr JM, et al. APOE E4 status predicts age-related cognitive decline in the ninth decade: longitudinal follow-up of the Lothian Birth Cohort 1921. Molecular psychiatry. 2012;17:315–24. doi: 10.1038/mp.2010.137. [DOI] [PubMed] [Google Scholar]
- 95.Holland D, McEvoy LK, Desikan RS, Dale AM. Enrichment and stratification for predementia Alzheimer disease clinical trials. PloS one. 2012;7:e47739. doi: 10.1371/journal.pone.0047739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–69. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 97.Honea RA, Cruchaga C, Perea RD, Saykin AJ, Burns JM, Weinberger DR, et al. Characterizing the role of brain derived neurotrophic factor genetic variation in Alzheimer’s disease neurodegeneration. PloS one. 2013;8:e76001. doi: 10.1371/journal.pone.0076001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lim YY, Villemagne VL, Laws SM, Pietrzak RH, Snyder PJ, Ames D, et al. APOE and BDNF polymorphisms moderate amyloid beta-related cognitive decline in preclinical Alzheimer’s disease. Molecular psychiatry. 2014 doi: 10.1038/mp.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Corneveaux JJ, Liang WS, Reiman EM, Webster JA, Myers AJ, Zismann VL, et al. Evidence for an association between KIBRA and late-onset Alzheimer’s disease. Neurobiology of aging. 2010;31:901–9. doi: 10.1016/j.neurobiolaging.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Milnik A, Heck A, Vogler C, Heinze HJ, de Quervain DJ, Papassotiropoulos A. Association of KIBRA with episodic and working memory: a meta-analysis. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2012;159B:958–69. doi: 10.1002/ajmg.b.32101. [DOI] [PubMed] [Google Scholar]
- 101.Need AC, Attix DK, McEvoy JM, Cirulli ET, Linney KN, Wagoner AP, et al. Failure to replicate effect of Kibra on human memory in two large cohorts of European origin. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2008;147B:667–8. doi: 10.1002/ajmg.b.30658. [DOI] [PubMed] [Google Scholar]
- 102.Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, et al. Common Kibra alleles are associated with human memory performance. Science. 2006;314:475–8. doi: 10.1126/science.1129837. [DOI] [PubMed] [Google Scholar]
- 103.Ritchie MD, Holzinger ER, Li R, Pendergrass SA, Kim D. Methods of integrating data to uncover genotype-phenotype interactions. Nature reviews Genetics. 2015;16:85–97. doi: 10.1038/nrg3868. [DOI] [PubMed] [Google Scholar]
- 104.Trzepacz PT, Yu P, Sun J, Schuh K, Case M, Witte MM, et al. Comparison of neuroimaging modalities for the prediction of conversion from mild cognitive impairment to Alzheimer’s dementia. Neurobiology of aging. 2014;35:143–51. doi: 10.1016/j.neurobiolaging.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 105.Zhang Z, Huang H, Shen D Alzheimer’s Disease Neuroimaging I. Integrative analysis of multi-dimensional imaging genomics data for Alzheimer’s disease prediction. Frontiers in aging neuroscience. 2014;6:260. doi: 10.3389/fnagi.2014.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bettens K, Sleegers K, Van Broeckhoven C. Genetic insights in Alzheimer’s disease. The Lancet Neurology. 2013;12:92–104. doi: 10.1016/S1474-4422(12)70259-4. [DOI] [PubMed] [Google Scholar]
- 107.Beadle GW, Tatum EL. Genetic Control of Biochemical Reactions in Neurospora. Proceedings of the National Academy of Sciences of the United States of America. 1941;27:499–506. doi: 10.1073/pnas.27.11.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144:986–98. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ala-Korpela M, Kangas AJ, Inouye M. Genome-wide association studies and systems biology: together at last. Trends in genetics: TIG. 2011;27:493–8. doi: 10.1016/j.tig.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 110.Califano A, Butte AJ, Friend S, Ideker T, Schadt E. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nature genetics. 2012;44:841–7. doi: 10.1038/ng.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hood L. Systems biology and p4 medicine: past, present, and future. Rambam Maimonides Med J. 2013;4:e0012. doi: 10.5041/RMMJ.10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nature reviews Genetics. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nature genetics. 2011;43:429–35. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nature genetics. 2011;43:436–41. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Walker DG, Whetzel AM, Serrano G, Sue LI, Beach TG, Lue LF. Association of CD33 polymorphism rs3865444 with Alzheimer’s disease pathology and CD33 expression in human cerebral cortex. Neurobiology of aging. 2015;36:571–82. doi: 10.1016/j.neurobiolaging.2014.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Malik M, Chiles J, 3rd, Xi HS, Medway C, Simpson J, Potluri S, et al. Genetics of CD33 in Alzheimer’s disease and acute myeloid leukemia. Human molecular genetics. 2015 doi: 10.1093/hmg/ddv092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zuk O, Schaffner SF, Samocha K, Do R, Hechter E, Kathiresan S, et al. Searching for missing heritability: designing rare variant association studies. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E455–64. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rhinn H, Fujita R, Qiang L, Cheng R, Lee JH, Abeliovich A. Integrative genomics identifies APOE epsilon4 effectors in Alzheimer’s disease. Nature. 2013;500:45–50. doi: 10.1038/nature12415. [DOI] [PubMed] [Google Scholar]
- 119.Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–20. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Penrod NM, Cowper-Sal-lari R, Moore JH. Systems genetics for drug target discovery. Trends in pharmacological sciences. 2011;32:623–30. doi: 10.1016/j.tips.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Janssens AC, van Duijn CM. Genome-based prediction of common diseases: advances and prospects. Human molecular genetics. 2008;17:R166–73. doi: 10.1093/hmg/ddn250. [DOI] [PubMed] [Google Scholar]
- 122.Fan M, Liu B, Zhou Y, Zhen X, Xu C, Jiang T, et al. Cortical thickness is associated with different apolipoprotein E genotypes in healthy elderly adults. Neuroscience letters. 2010;479:332–6. doi: 10.1016/j.neulet.2010.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Naj AC, Jun G, Reitz C, Kunkle BW, Perry W, Park YS, et al. Effects of Multiple Genetic Loci on Age at Onset in Late-Onset Alzheimer Disease A Genome-Wide Association Study. JAMA neurology. 2014;71:1394–404. doi: 10.1001/jamaneurol.2014.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Doody RS, Thomas RG, Farlow M, Iwatsubo T, Vellas B, Joffe S, et al. Phase 3 trials of solanezumab for mild-to-moderate Alzheimer’s disease. The New England journal of medicine. 2014;370:311–21. doi: 10.1056/NEJMoa1312889. [DOI] [PubMed] [Google Scholar]
- 125.Salloway S, Sperling R, Fox NC, Blennow K, Klunk W, Raskind M, et al. Two Phase 3 Trials of Bapineuzumab in Mild-to-Moderate Alzheimer’s Disease. New Engl J Med. 2014;370:322–33. doi: 10.1056/NEJMoa1304839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sabuncu MR, Buckner RL, Smoller JW, Lee PH, Fischl B, Sperling RA, et al. The association between a polygenic Alzheimer score and cortical thickness in clinically normal subjects. Cereb Cortex. 2012;22:2653–61. doi: 10.1093/cercor/bhr348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rodriguez-Rodriguez E, Sanchez-Juan P, Vazquez-Higuera JL, Mateo I, Pozueta A, Berciano J, et al. Genetic risk score predicting accelerated progression from mild cognitive impairment to Alzheimer’s disease. J Neural Transm. 2013;120:807–12. doi: 10.1007/s00702-012-0920-x. [DOI] [PubMed] [Google Scholar]
- 128.Garraway LA. Genomics-Driven Oncology: Framework for an Emerging Paradigm. J Clin Oncol. 2013;31:1806–14. doi: 10.1200/JCO.2012.46.8934. [DOI] [PubMed] [Google Scholar]
- 129.Van Allen EM, Wagle N, Levy MA. Clinical Analysis and Interpretation of Cancer Genome Data. J Clin Oncol. 2013;31:1825–33. doi: 10.1200/JCO.2013.48.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cook D, Brown D, Alexander R, March R, Morgan P, Satterthwaite G, et al. Lessons learned from the fate of AstraZeneca’s drug pipeline: a five-dimensional framework. Nat Rev Drug Discov. 2014;13:419–31. doi: 10.1038/nrd4309. [DOI] [PubMed] [Google Scholar]
- 131.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat Rev Drug Discov. 2013;12:581–94. doi: 10.1038/nrd4051. [DOI] [PubMed] [Google Scholar]
- 132.Burgess S, Butterworth A, Malarstig A, Thompson SG. Use of Mendelian randomisation to assess potential benefit of clinical intervention. Bmj. 2012;345:e7325. doi: 10.1136/bmj.e7325. [DOI] [PubMed] [Google Scholar]
- 133.International Genomics of Alzheimer’s Disease C. Convergent genetic and expression data implicate immunity in Alzheimer’s disease. Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2014 doi: 10.1016/j.jalz.2014.05.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Heneka MT, Golenbock DT, Latz E. Innate immunity in Alzheimer’s disease. Nature immunology. 2015;16:229–36. doi: 10.1038/ni.3102. [DOI] [PubMed] [Google Scholar]
- 135.Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, et al. TREM2 variants in Alzheimer’s disease. The New England journal of medicine. 2013;368:117–27. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yu J, Thomson JA. Pluripotent stem cell lines. Genes & development. 2008;22:1987–97. doi: 10.1101/gad.1689808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yagi T, Ito D, Okada Y, Akamatsu W, Nihei Y, Yoshizaki T, et al. Modeling familial Alzheimer’s disease with induced pluripotent stem cells. Human molecular genetics. 2011;20:4530–9. doi: 10.1093/hmg/ddr394. [DOI] [PubMed] [Google Scholar]
- 138.Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482:216–20. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Barrett R, Ornelas L, Yeager N, Mandefro B, Sahabian A, Lenaeus L, et al. Reliable generation of induced pluripotent stem cells from human lymphoblastoid cell lines. Stem cells translational medicine. 2014;3:1429–34. doi: 10.5966/sctm.2014-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Young JE, Boulanger-Weill J, Williams DA, Woodruff G, Buen F, Revilla AC, et al. Elucidating Molecular Phenotypes Caused by the SORL1 Alzheimer’s Disease Genetic Risk Factor Using Human Induced Pluripotent Stem Cells. Cell stem cell. 2015 doi: 10.1016/j.stem.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D’Avanzo C, et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature. 2014;515:274–8. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wolf SM, Lawrenz FP, Nelson CA, Kahn JP, Cho MK, Clayton EW, et al. Managing incidental findings in human subjects research: analysis and recommendations. J Law Med Ethics. 2008;36:219–48. 1. doi: 10.1111/j.1748-720X.2008.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Weiner C. Anticipate and communicate: ethical management of incidental and secondary findings in the clinical, research, and direct-to-consumer contexts (December 2013 report of the Presidential Commission for the Study of Bioethical Issues) Am J Epidemiol. 2014;180:562–4. doi: 10.1093/aje/kwu217. [DOI] [PubMed] [Google Scholar]
- 144.Morello-Frosch R, Varshavsky J, Liboiron M, Brown P, Brody JG. Communicating results in post-Belmont era biomonitoring studies: lessons from genetics and neuroimaging research. Environ Res. 2015;136:363–72. doi: 10.1016/j.envres.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Booth TC, Jackson A, Wardlaw JM, Taylor SA, Waldman AD. Incidental findings found in “healthy” volunteers during imaging performed for research: current legal and ethical implications. Br J Radiol. 2010;83:456–65. doi: 10.1259/bjr/15877332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shoemaker JM, Holdsworth MT, Aine C, Calhoun VD, de La Garza R, Feldstein Ewing SW, et al. A practical approach to incidental findings in neuroimaging research. Neurology. 2011;77:2123–7. doi: 10.1212/WNL.0b013e31823d7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Booth TC, Waldman AD, Wardlaw JM, Taylor SA, Jackson A. Management of incidental findings during imaging research in “healthy” volunteers: current UK practice. Br J Radiol. 2012;85:11–21. doi: 10.1259/bjr/73283917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jarvik GP, Amendola LM, Berg JS, Brothers K, Clayton EW, Chung W, et al. Return of genomic results to research participants: the floor, the ceiling, and the choices in between. Am J Hum Genet. 2014;94:818–26. doi: 10.1016/j.ajhg.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Berg JS, Amendola LM, Eng C, Van Allen E, Gray SW, Wagle N, et al. Processes and preliminary outputs for identification of actionable genes as incidental findings in genomic sequence data in the Clinical Sequencing Exploratory Research Consortium. Genet Med. 2013;15:860–7. doi: 10.1038/gim.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wolf SM, Crock BN, Van Ness B, Lawrenz F, Kahn JP, Beskow LM, et al. Managing incidental findings and research results in genomic research involving biobanks and archived datasets. Genet Med. 2012;14:361–84. doi: 10.1038/gim.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Suhr J, Gunstad J. ‘Diagnosis threat’: the effect of negative expectations on cognitive performance in head injury. J Clin Exp Neuropsychol. 2002;24:448–57. doi: 10.1076/jcen.24.4.448.1039. [DOI] [PubMed] [Google Scholar]
- 152.Lineweaver T, Bondi M, Galasko D, Salmon D. Knowledge of APOE genotype affects subjective and objective memory performance in healthy older adults. Am J Psychiatry. 2013;171:201–8. doi: 10.1176/appi.ajp.2013.12121590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shalowitz DI, Miller FG. Communicating the results of clinical research to participants: Attitudes, practices, and future directions. PLoS medicine. 2008:5. doi: 10.1371/journal.pmed.0050091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Neumann P, Cohen J, Hammitt J. Willingness-to-pay for predictive tests with no immediate treatment implications: a survey of US residents. Health Econ. 2012;21:238–51. doi: 10.1002/hec.1704. [DOI] [PubMed] [Google Scholar]
- 155.Wikler E, Blendon R, Benson J. Would you want to know? Public attitudes on early diagnostic testing for alzheimer’s disease. Alzheimers Res Ther. 2013;5:43. doi: 10.1186/alzrt206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Klein EP, Kaye J. Dementia specialists and early adoption of amyloid imaging. J Alzheimers Dis. 2013;33:445–50. doi: 10.3233/JAD-2012-121216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Johnson K, Minoshima S, Bohnen N, Donohoe K, Foster N, Herscovitch P, et al. Appropriate use criteria for amyloid PET: a report of the amyloid imaging task force, the society of nuclear medicine and molecular imaging, and the alzheimer’s association. J Nucl Med. 2013;54:14. doi: 10.2967/jnumed.113.120618. [DOI] [PubMed] [Google Scholar]
- 158.Johnson K, Minoshima S, Bohnen N, Donohoe K, Foster N, Herscovitch P, et al. Update on appropriate use criteria for amyloid PET imaging: dementia experts, mild cognitive impairment and education. Alzheimers Dement. 2013;9:e106–9. doi: 10.1016/j.jalz.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 159.National Coverage Analysis. Proposed decision memo for beta amyloid positron emission tomography in dementia and neurodegenerative disease (CAG-00431N) 2013 [Google Scholar]
- 160.Rogers MB. Coverage denial for amyloid scans riles Alzheimer’s community. AlzForum; 2013 [Google Scholar]
- 161.Shulman M, Harkins K, Green RC, Karlawish J. Using AD biomarker research results for clinical care: A survey of ADNI investigators. Neurology. 2013 doi: 10.1212/WNL.0b013e3182a55f4a. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Dressler LG, Smolek S, Ponsaran R, Markey JM, Starks H, Gerson N, et al. IRB perspectives on the return of individual results from genomic research. Genet Med. 2012;14:215–22. doi: 10.1038/gim.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Burke W, Evans BJ, Jarvik GP. Return of results: ethical and legal distinctions between research and clinical care. Am J Med Genet C Semin Med Genet. 2014;166C:105–11. doi: 10.1002/ajmg.c.31393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fabsitz RR, McGuire A, Sharp RR, Puggal M, Beskow LM, Biesecker LG, et al. Ethical and practical guidelines for reporting genetic research results to study participants: updated guidelines from a National Heart, Lung, and Blood Institute working group. Circ Cardiovasc Genet. 2010;3:574–80. doi: 10.1161/CIRCGENETICS.110.958827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Biesecker LG, Green RC. Diagnostic clinical genome and exome sequencing. N Engl J Med. 2014;370:2418–25. doi: 10.1056/NEJMra1312543. [DOI] [PubMed] [Google Scholar]
- 166.Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–9. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–74. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.ACMG Board of Directors. ACMG policy statement: Updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet Med. 2015;17:68–9. doi: 10.1038/gim.2014.151. [DOI] [PubMed] [Google Scholar]
- 169.Green RC, Lupski J, Biesecker LG. Reporting genomic sequencing results to ordering clinicians: incidental, but not exceptional. JAMA. 2013;310:365–6. doi: 10.1001/jama.2013.41703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wolf SM, Branum R, Koenig BA, Petersen GM, Berry SA, Beskow LM, et al. Returning a research participant’s genomic results to relatives: analysis and recommendations. J Law Med Ethics. doi: 10.1111/jlme.12288. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Green RC, Roberts JS, Cupples LA, Relkin NR, Whitehouse PJ, Brown T, et al. Disclosure of APOE genotype for risk of Alzheimer’s disease. N Engl J Med. 2009;361:245–54. doi: 10.1056/NEJMoa0809578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Roberts J, Christensen K, Green RC. Using Alzheimer’s disease as a model for genetic risk disclosure: implications for personal genomics. Clin Genet. 2011;80:407–14. doi: 10.1111/j.1399-0004.2011.01739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lautenbach DM, Christensen KD, Sparks JA, Green RC. Communicating genetic risk information for non-Mendelian variants. Annu Rev Genomomics Hum Genet. 2013;14:491–513. doi: 10.1146/annurev-genom-092010-110722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Green RC, Christensen KD, Cupples LA, Relkin NR, Whitehouse PJ, Royal CD, et al. A randomized noninferiority trial of condensed protocols for genetic risk disclosure of Alzheimer’s disease. Alzheimers Dement. 2014 doi: 10.1016/j.jalz.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gymrek M, McGuire A, Golan D, Halperin E, Erlich Y. Identifying personal genomes by surname inference. Science. 2012;339:321–4. doi: 10.1126/science.1229566. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.