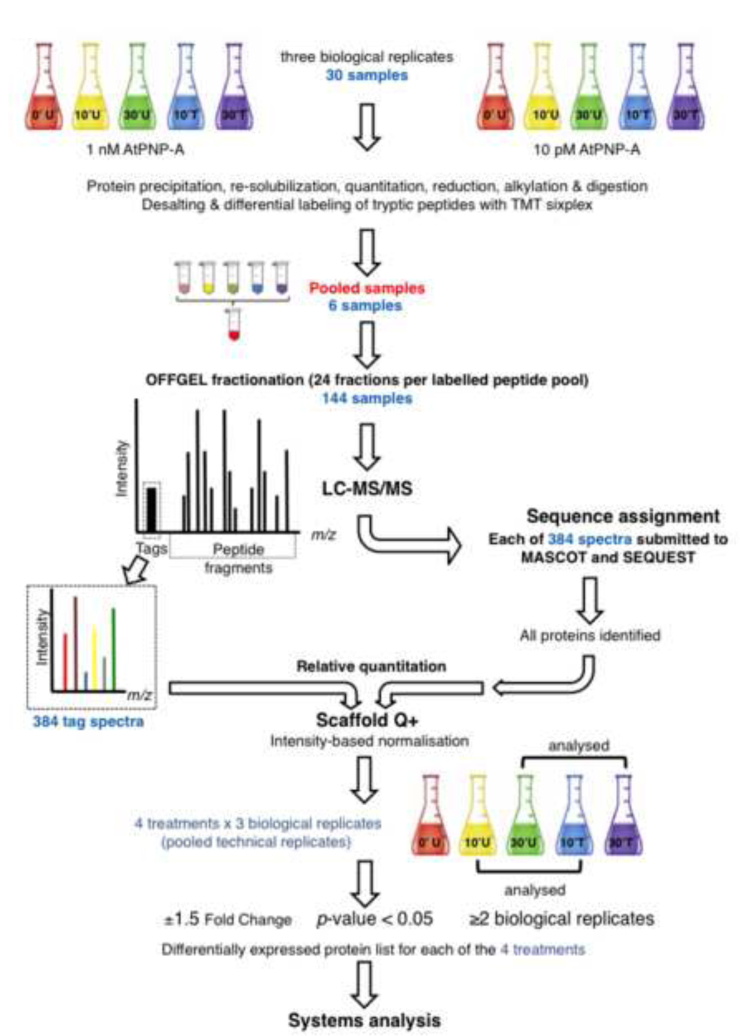
Fig. 1.
Overview of the experiment. Arabidopsis cell suspension cultures were treated (T) with AtPNP-A (1 nM or 10 pM) or water (mock, U) for 0, 10 or 30 min. Three biological replicates were performed for each, resulting in 30 samples. Protein extraction, quantitation, reduction, alkylation and digestion were performed for each samples followed by desalting before differential labeling of the tryptic peptides with TMT sixplex. AtPNP-A-treated cells collected at 10 and 30 min post-treatment were labeled with m/z 129 TMT and m/z 130 TMT while the 0, 10, 30 min mock treated cells were labeled with m/z 126 TMT, m/z 127 TMT and m/z 128 TMT, respectively. Equal amounts of labeled peptides from the corresponding biological replicates were then pooled to create 6 combined samples (3 biological replicates for 1 nM AtPNP-A treatments and 3 biological replicates for 10 pM AtPNP-A treatments). OFFGEL fractionation was performed giving rise to 24 fractions per labeled peptide pool. Each of these samples was analyzed by LC/MS–MS. Each spectrum was analyzed independently by MASCOT and SEQUEST for sequence assignment (Supplementary Tables 1 and 2). Scaffold Q+ was then used to relatively quantify all identified proteins. Differential protein expression was considered significant if the combined data from pooled technical replicates for a given biological replicate was greater or equal to |±1.5| of the related mock treatment, verified by Mann–Whitney test (p-value <0.05), in at least two out of three biological replicates (Tables 1 and 2). Differentially expressed proteins were then examined by GO and gene expression analysis.