Abstract
This data article reports the establishment of the first pan-transcriptome resources for the Brassica A and C genomes. These were developed using existing coding DNA sequence (CDS) gene models from the now-published Brassica oleracea TO1000 and Brassica napus Darmor-bzh genome sequence assemblies representing the chromosomes of these species, along with preliminary CDS models from an updated Brassica rapa Chiifu genome sequence assembly. The B. rapa genome sequence scaffolds required splitting and re-ordering to match the expected genome organisation based on a high density SNP linkage map, but the B. oleracea assembly was used unchanged. The resulting B. rapa (A genome) pseudomolecules contained 47,656 ordered CDS models and the B. oleracea (C genome) pseudomolecules contained 54,766 ordered CDS models. Interpolation of B. napus CDS models not already represented by orthologues resulted in 52,790 and 63,308 ordered CDS models in the A and C pan-transcriptomes, an increase of 13,676 overall. Comparison of the organisation of this resource with publicly available genome sequences for B. napus showed excellent consistency for the B. napus Darmor-bzh resource, but more breakdown of collinearity for the B. napus ZS11 resource. CDS datasets comprising the pan-transcriptomes are available with this article (B. rapa) or from public repositories (B. oleracea and B. napus).
Specifications Table
| Subject area | Biology |
| More specific subject area | Plant genome organisation |
| Type of data | CDS gene model sequences for the A genome, in FASTA format. Tables (in the form of MS Excel spreadsheets) providing A genome pseudomolecule specification based on genome sequence scaffolds, inferred order and anchoring positions in the A and C genome pseudomolecules for CDS models and a figure illustrating the collinearity of the ordered pan-transcriptome and two genome sequences reported for B. napus. |
| How data was acquired | CDS gene model sequences for the A genome were developed as part of the reported work. Genome sequence scaffolds and other CDS data were obtained from the groups generating them prior to publication. |
| Data format | The data accompanying this article are provided as text files (for B. rapa CDS models and R scripts) and MS Excel spreadsheets providing CDS and scaffold identifiers and sequence similarity coordinates. |
| Experimental factors | n/a |
| Experimental features | CDS modelling was undertaken using V2.0 B. rapa genome sequence scaffolds. A previously-reported set of Brassica A genome pseudomolecules was used to produce improved pseudomolecules derived from an updated B. rapa genome assembly in order to represent the organisation of the A genome in B. napus. Integration and interpolation of gene models called only in a B. napus genome sequence was undertaken, resulting in the establishment of a pan-transcriptome resource for the Brassica A and C genomes. Collinearity analysis with public B. napus genome sequences was undertaken, based on BLAST similarity hits of CDS models, to compare the order of genes in the pan-transcriptome resource with that of their orthologues in two published B. napus genome sequences. |
| Data source location | SRA, NCBI, ENA |
| Data accessibility | All genome sequence datasets were provided for analysis prior to publications, but are now available: |
| The B. napus Darmor-bzh assembly is available at ENA (European Nucleotide Archive), in the WGS section for contigs (accession numbers CCCW010000001 to CCCW010044187) and the CON section for scaffolds, chromosomes, and annotation (accession numbers LK031787 to LK052685). The B. napus ZS11 assembly is available at http://www.ncbi.nlm.nih.gov/Traces/wgs/?val=JMKK01#. The B. oleracea assembly is available via Sequence Read Archive accession number PRJNA158027. The B. rapa version 2 assembly is in the process of publication and in the meantime is available from Xiaowu Wang (wangxiaowu@caas.cn). |
Value of the data
-
•
Provides an updated pseudomolecule description, with genome sequence scaffolds from B. rapa and B. oleracea representing genome organisation in B. napus
-
•
Provides for the first time pan-transcriptome resources for use in Brassica species containing the A and/or C genomes
-
•
Provides insights into the extent of gene content variation between the Brassica A and C genomes as represented in an allopolyploid and its diploid progenitors
-
•
Provides a hypothetical gene order resource for the Brassica A and C pan-genomes for use in genome evolution studies and Associative Transcriptomics.
1. Experimental design, materials and methods
Transcriptome-based molecular marker systems have been developed and deployed with great success in the crop species B. napus for both genome organisation studies [1] and association genetics [2]. These studies exploit mRNAseq data, which need to be mapped to a suitable transcriptome reference sequence for single nucleotide polymorphism (SNP) identification and transcript quantification. The first generation approach used unigene assemblies as the reference sequences [3], which permitted some resolution of the contributions to the transcriptome of homoeologous gene pairs [4]. However, the genome sequences reported for B. napus Darmor-bzh indicate that sequence exchanges between the constituent genomes of this allotetraploid species (A genome from an unknown B. rapa and C genome from an unknown B. oleacea) may occur very frequently [5], making it imperative that the genome-of-origin of any given gene be determined as clearly as possible. The most reliable way of achieving this is to base resources primarily on those derived from the constituent genomes in the diploid progenitors of B. napus, i.e. from B. rapa and B. oleracea. As an improvement on the existing resource based on unigenes assembled across Brassica species [3,6], we therefore aimed to develop a new transcriptome reference, based on coding DNA sequence (CDS) gene models derived primarily from the Brassica A and C genomes as represented in the progenitor species. As the B. napus genome sequence annotation identified many gene models without orthologues in B. rapa and B. oleracea, we further aimed to interpolate those B. napus-specific CDS models, thus producing pan-transcriptome resources for the Brassica A and C genomes as represented by the union of orthologous genes of B. rapa, B. oleracea and B. napus.
The version 2 B. rapa Chiifu genome sequence scaffolds represent a major advance on the published version 1 sequences [7] in that they provide more comprehensive coverage of the genome, with aggregate scaffold size increasing from 248 Mb to 370 Mb. A preliminary annotation was undertaken of the genome sequence scaffolds that had been organised into chromosomes, essentially as described for the version 1 genome sequences [7]. Briefly: Genscan and Augustus with parameters established using Arabidopsis thaliana gene models were used to perform de novo gene predictions in the new genome assembly of B. rapa, after masking the Class I and Class II transposable elements. The predicted genes with CDS models shorter than 150 bp were filtered out. We further performed homology based gene prediction by aligning A. thaliana, Carica papaya, Populus trichocarpa, Vitis vinifera and Oryza sativa protein sequences to the B. rapa genome. TBLASTN was used to do fast alignment (threshold e-value 1E−5), then Genewise was used to do precise alignment. Additionally, we assembled the Brassica ESTs downloaded from NCBI using PASA and aligned them to B. rapa genome by BLAT. Considering that the fragmented exons in EST data might lead to false results, we filtered out alignments with gaps (introns) that span over 10 kb in length. We then ran GLEAN to merge the gene sets generated from de novo and homology-based predictions, using mRNA-Seq data as the supporting evidence. Finally, the B. rapa gene set was aligned to the TE protein database of Repbase, those hits with e-value>1E−5 and coverage≥50% were filtered out. The remaining gene models were reported as Brassica gene set Version 2.0 (Additional file 1). These CDS models were then used in sequence similarity searches using BLAST to identify the highest-scoring significant hit (threshold e-value 1E−30) for each CDS model in both the version 2 B. rapa Chiifu genome sequence scaffolds and the A genome pseudomolecules reported previously [6], based on the version 1 B. rapa Chiifu genome sequence [7] that had been reordered relative to the B. napus genome via high density transcriptome SNP linkage mapping [1]. This enabled the identification of chimeric scaffolds in the version 2 assembly that could be split (Additional file 2) and re-organised (Additional file 3) to form pseudomolecules representative of the organisation of the Brassica A genome. The CDS models from the B. oleracea TO1000 [8] genome sequence were similarly used to assess collinearity with the C genome pseudomolecules reported previously [6] and were found to be in excellent agreement, so the B. oleracea TO1000 assembly was adopted unaltered as representing the Brassica C genome pseudomolecule resource.
The B. rapa Chiifu CDS, along with CDS from the published B. oleracea TO1000 genome sequence [8], was mapped onto the respective genome sequence pseudomolecules using BLAST to identify the highest-scoring significant hit (threshold e-value 1E−30). This resulted in the mapping and ordering of 47,656 B. rapa CDS models to the A genome and 54,766 B. oleracea CDS models to the C genome. A total of 101,040 CDS models were annotated in the B. napus Darmor-bzh genome [5]. Of these, 80,927 CDS models which had been anchored to the 19 B. napus pseudomolecules were mapped onto the respective (B. rapa and B. oleracea-based) genome sequence pseudomolecules by BLAST (threshold e-value 1E−30). B. napus CDS models mapping redundantly with CDS models derived from B. rapa and B. oleracea (threshold e-value 1E−30) were excluded, resulting in the addition of 2165 and 3032 CDS models to the A and C genomes, respectively. Finally, CDS models from the B. napus Darmor-bzh genome sequence that did not have significant (threshold e-value 1E−30) BLAST hits in the (B. rapa and B. oleracea-based) genome sequence pseudomolecules were interpolated based on the positions of flanking gene models that did map. This was done by combining the B. napus Darmor-bzh CDS models׳ sorted location on the B. napus Darmor-bzh chromosome with the mapped location of flanking genes on the B. rapa or B. oleracea-based pseudomolecules using an R script (Additional file 4) to perform the following: (1) Sort B. napus CDS models by B. napus Darmor-bzh pseudomolecules, then by their B. rapa or B. oleracea-based pseudomolecules hit locations then (2) CDS models (or runs of adjacent CDS models) that do not have a hit onto the B. rapa or B. oleracea-based pseudomolecules are interpolated onto those pseudomolecules with a three digit suffix starting from the boundary of the point of insertion. When the boundaries are not in the right order, the interpolation starts from the closest boundary number to the mean of the nearest 10 neighbours of the run of CDS models. If there is no mapping in the 10 nearest neighbours, the interpolation starts from the minimum of the boundary numbers. This resulted in the addition of 2969 and 5510 further CDS models to the A and C genomes, respectively. The final AC pan-transcriptome resource therefore comprises a total of 116,098 hypothetically ordered CDS models (Additional file 5,6,7), 52,790 in the Brassica A genome and 63,308 in the Brassica C genome. This represents an increase of 35,171 over the 80,927 CDS models annotated in the published B. napus Darmor-bzh pseudomolecules, 15,058 over the complement of gene models for B. napus including the 20,113 in sequence scaffolds not incorporated into the B. napus Darmor-bzh pseudomolecules [5] and 13,676 more than had been identified in the B. rapa and B. oleracea pseudomolecules.
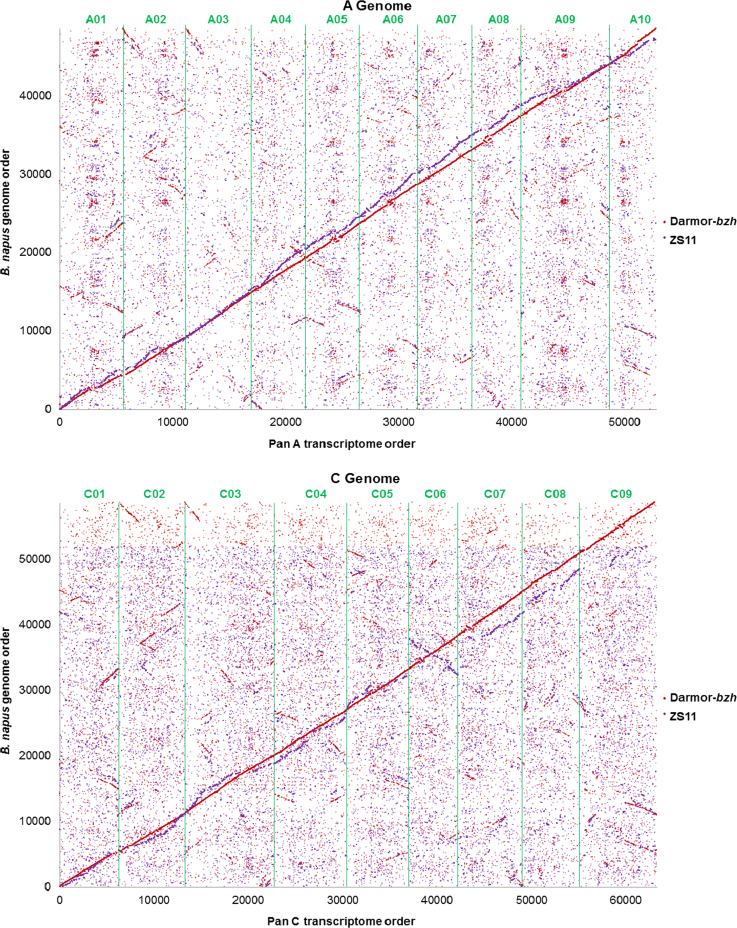
The order of CDS models in the pan transcriptome was compared with the order of orthologous sequences in two publicly-available B. napus genome sequence resources. This was conducted by sequence similarity search using BLAST to identify the highest-scoring significant hit (threshold e-value 1E−30) for each CDS model in the pan-transcriptome in each of the B. napus Darmor-bzh and B. napus ZS11 chromosome assemblies. Of the aggregate 116,098 CDS models in the pan-transcriptome, 107,292 (92.4%) returned significant hits (threshold e-value 1E−30) in the B. napus Darmor-bzh assembly and 99,395 (85.6%) returned significant hits in the B. napus ZS11 assembly. The order of these best similarity matches in each resource is illustrated in Fig. 1. The inferred gene order in the pan-transcriptome and the B. napus Darmor-bzh genome assembly shows excellent collinearity. A small number of local rearrangements can be observed in regions with relative high densities of non-collinear matches, possibly corresponding to paracentromeric regions. In addition, two prominent segments shadowing the main collinearity diagonal can be observed amongst the background of CDS models mapping to non-orthologous positions. Such shadows have been observed in previous studies [6] and were shown to correspond to sequences missing from the genome sequence resource, with consequent mapping of sequences to one of the two paralogous segments of these paleohexaploid genomes. The inferred gene order in the pan-transcriptome and the B. napus ZS11 genome assembly show extensive collinearity, but with more disruption by rearrangements than was observed with B. napus Darmor-bzh resource. Linkage group C6 is also presented in the B. napus ZS11 genome sequence resource in the opposite orientation to the current reference genetic map for the Brassica C genome. These analyses, which together indicate extensive collinearity of the Brassica A and C genomes as represented in the allotetraploid B. napus and representatives of its progenitors, are also consistent with early observations of extensive collinearity, but with some divergence in gene content between orthologous regions of Brassica genomes, including both loss and mobility of coding sequences [9].
Fig. 1.
Collinearity of ordered pan-transcriptomes and the genome sequences of B. napus Darmor-bzh and B. napus ZS11. The positions of best sequence matches in the B. napus chromosome assemblies are plotted for CDS models with significant similarity matches (threshold e-value 1E−30) in the B. napus Darmor-bzh assembly and B. napus ZS11 assembly.
Acknowledgements
This work was supported by UK Biotechnology and Biological Sciences Research Council (BBSRC BB/L027844/1, BB/L002124/1), National Natural Science Foundation of China (Grant no. 31471536), the 973 Program 2012CB113900 to XW and FC; the 863 Program 2012AA100101 to XW; the National Natural Science Foundation of China NSFC Grant 31301771 to FC. Research was conducted in the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, P.R. China.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2015.06.016.
Supplementary materials
Supplementary data
References
- 1.Bancroft I., Morgan C., Fraser F., Higgins J., Wells R., Clissold L., Baker D., Long Y., Meng J., Wang X. Dissecting the genome of the polyploid crop oilseed rape by transcriptome sequencing. Nat. Biotechnol. 2011;29:762–766. doi: 10.1038/nbt.1926. [DOI] [PubMed] [Google Scholar]
- 2.Harper A.L., Trick M., Higgins J., Fraser F., Clissold L., Wells R., Hattori C., Werner P., Bancroft I. Associative transcriptomics of traits in the polyploid crop species Brassica napus. Nat. Biotechnol. 2012;30:798–802. doi: 10.1038/nbt.2302. [DOI] [PubMed] [Google Scholar]
- 3.Trick M., Long Y., Meng J., Bancroft I. Single nucleotide polymorphism (SNP) discovery in the polyploid Brassica napus using Solexa transcriptome sequencing. Plant Biotechnol. J. 2009;7:334–346. doi: 10.1111/j.1467-7652.2008.00396.x. [DOI] [PubMed] [Google Scholar]
- 4.Higgins J., Magusin A., Trick M., Fraser F., Bancroft I. Use of mRNA-seq to discriminate contributions to the transcriptome from the constituent genomes of the polyploidy crop species Brassica napus. BMC Genomics. 2012;13:247. doi: 10.1186/1471-2164-13-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chalhoub B. Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science. 2014;345:950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 6.Bancroft I., Fraser F., Morgan C., Trick M. Collinearity analysis of Brassica A and C genomes based on an updated inferred unigene order. Data Br. 2015;3:51–55. doi: 10.1016/j.dib.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X. The genome of the mesohexaploid crop species Brassica rapa. Nat. Genet. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 8.Parkin I. Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol. 2014;15:R77. doi: 10.1186/gb-2014-15-6-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung F., Trick M., Drou N., Lim Y.P., Park J.-Y., Kwon S.-J., Kim J.-A., Scott R., Pires J.C., Paterson A.H. Comparative analysis between homoeologous genome segments of Brassica napus and its progenitor species reveals extensive sequence-level divergence. Plant Cell. 2009;21:1912–1928. doi: 10.1105/tpc.108.060376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data