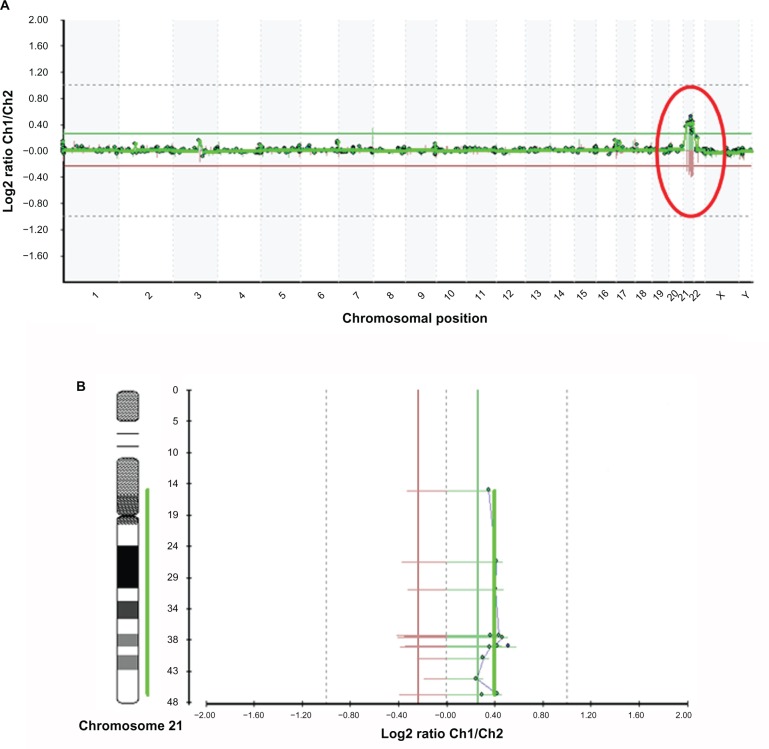
Figure 4.
A. Microarray profile for Down syndrome (47,XX+21) identified by microarray-based comparative genomic hybridization (array CGH) using GOLD Chip. Each clone represented on the array is arranged along the x-axis according to its location on the chromosome with the most distal/telomeric p-arm clones on the left and the most distal/telomeric q-arm clones on the right. The green lines represent the log2 ratios from the first experiment (patient Cy3/control Cy5), whereas the red plots represent the log2 ratios obtained from the second experiment in which the dyes have been reversed (patient Cy5/control Cy3). B. Microarray profiles of chromosome 21. The green line represents the patient-to-control fluorescence intensity ratios (gain of DNA); the red line represents dye-reversed control-to-patient fluorescence ratios (loss of DNA). Some clones appear in the same horizontal line because they are overlapping. In the image are shown only the 13 clones that, after data analysis and normalization, were selected by Bluefuse considering the parameters of the experiments. Some regions of chromosome 21 (p-arm) are not covered by BAC clones.