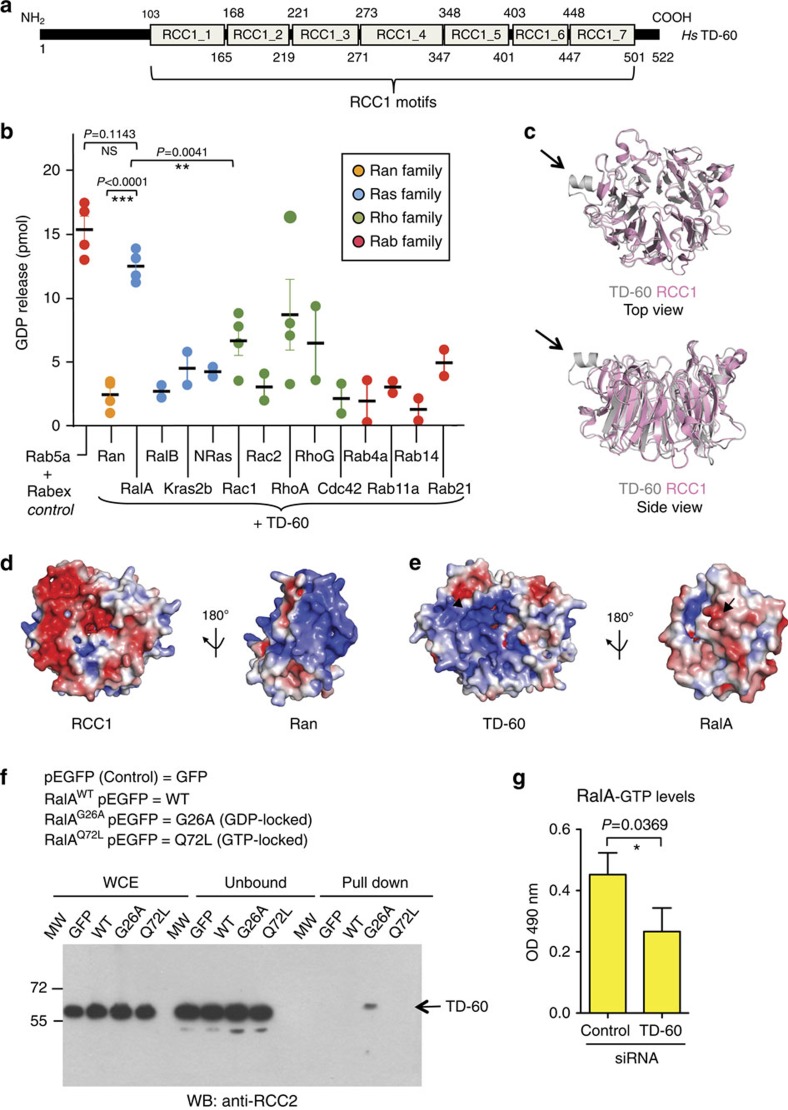
Figure 1. TD-60 has GEF activity towards RalA.
(a) Schematic diagram of the human TD-60 protein showing RCC1 motif distribution. (b) Human TD-60 was tested against a representative panel of human small GTPase proteins using a published GDP-release assay30,70. Nucleotide exchange was calculated as pmoles of GDP released (n=2,4). Graph reports mean±s.e.m. Statistical significance was determined using unpaired Student's t-test (***P<0.0001). Additional statistical values can be found in Supplementary Table 1 (c) Superposition of a TD-60 structure (grey) predicted using Phyre2 ( http://www.sbg.bio.ic.ac.uk/phyre2)34, onto the crystal structure of RCC1 (pink—PDB id: 1I2M) suggests that the overall structure of TD-60 is similar to RCC1 with an insertion of an α-helical segment (highlighted by arrows) which is unique to higher vertebrate TD-60. (d) Surface representation of RCC1 and Ran highlighting the charge complementarity of their binding interfaces (both shown facing the viewer, such that a 180° flip of one of the binding partners brings the two interacting surfaces together). Blue and red represent positive and negatively charged regions, respectively. Electrostatic surface potential of RCC1 and Ran (PDB id: 1I2M) was calculated using the APBS plug-in in PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC). (e) Similar analysis of surface charge for TD-60 and RalA (PDB id: 2A78). (f) Pull-down assay reveals that TD-60 interacts specifically with RalA when bound to GDP in cells. HeLa Kyoto cells were transfected to express GFP-tagged RalA WT, G26A (GDP) and Q72L (GTP), and endogenous TD-60 was pulled down using GFP-Trap-M beads. (g) TD-60 depletion reduces RalA activation in cells. HeLa cells were transfected with TD-60 siRNA and RalA-GTP levels in lysates were measured by G-LISA assay. Graph reports the mean absorbance at 490 nm±s.e.m. (n=3), with statistical significance assessed using unpaired Student's t-test (*P<0.05).