Figure 1. Homology modelling and topology of human CASD1.
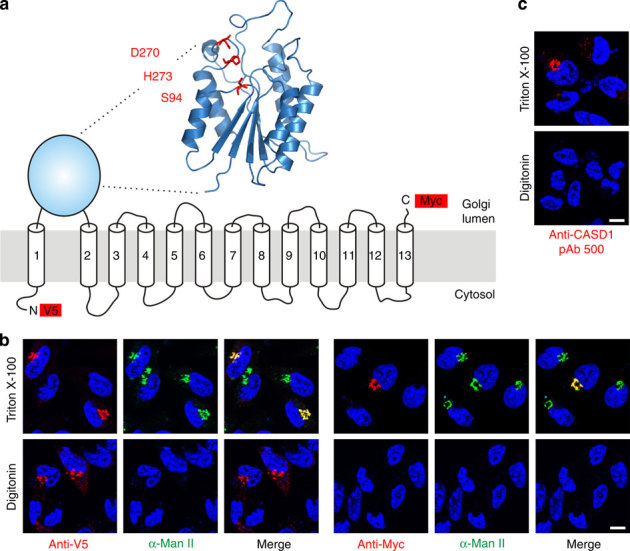
(a) Proposed model of CASD1. The overall architecture of the catalytic domain was predicted by homology modelling using the server Phyre2 and the crystal structure of an isoamyl acetate-hydrolysing esterase from Saccharomyces cerevisiae (PDB ID 3mil) as template. The model encompasses residues 83-290 of CASD1 and the predicted catalytic triad is highlighted in red. TMDs are depicted as predicted by the program TMHMM 2.0 (Supplementary Fig. 1). Epitope tags used for the localization of N- and C-terminus are shown as red boxes. (b) Orientation of the N- and C-terminus of CASD1. LM-TK− cells expressing CASD1 with an N-terminal V5-tag and a C-terminal Myc-tag (V5-CASD1-Myc) were fixed and permeabilized with Triton X-100 (0.2%) or digitonin (5 μg ml−1). Cells were stained with anti-V5 or anti-Myc mAb (red) in combination with anti-α-mannosidase II (α-Man II) pAb (green) recognizing the luminal domain of the Golgi marker α-Man II. Nuclei were counterstained with DAPI (blue). (c) Orientation of the catalytic domain of CASD1. LM-TK− cells expressing CASD1 were permeabilized as described in b and stained with pAb 500 directed against the catalytic domain of CASD1. Nuclei were counterstained with DAPI (blue). Scale bar, 10 μm. DAPI, 4,6-diamidino-2-phenylindole; TMD, transmembrane domain.
