Figure 1. E2a is required for subsets of Smad2/3 binding, and for global patterns of Smad2/3 target gene expression.
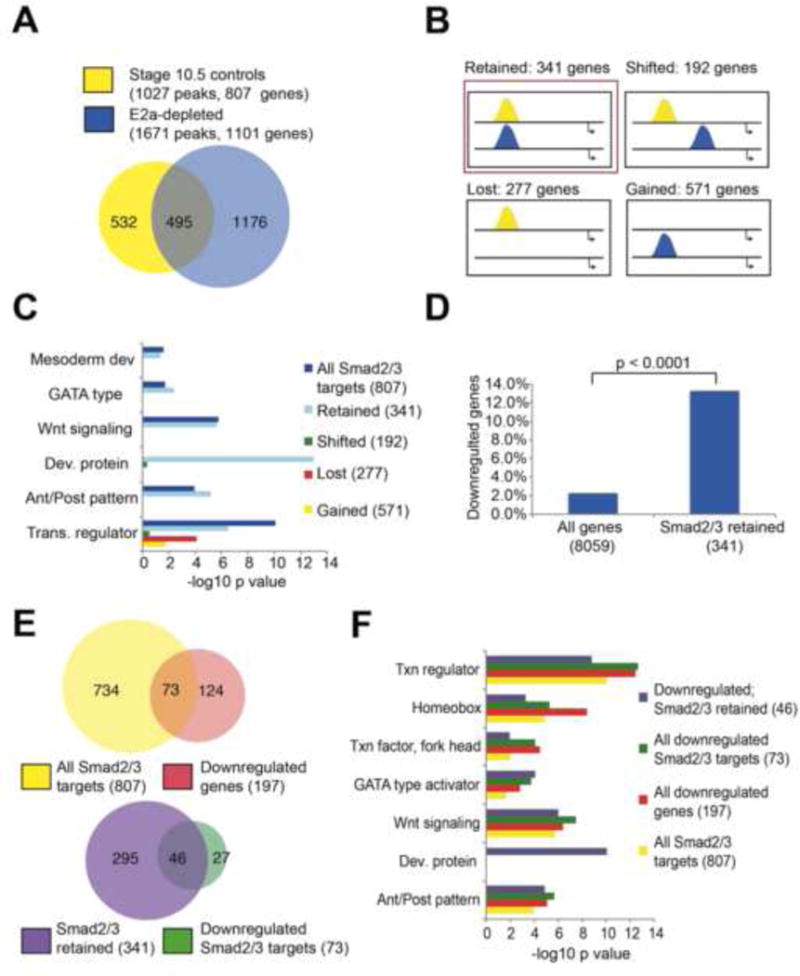
A) Smad2/3 targets 1027 distinct genomic regions in control stage 10.5 embryos (yellow), and 1671 regions in E2a-depleted embryos (blue). 495 regions are targeted in both conditions. B) Categories of Smad2/3 binding behavior in E2a-depleted embryos (blue) relative to controls (yellow). C) DAVID clustering analysis shows enrichment for developmental terms in all Smad2/3 associated genes, and for genes that have stable Smad2/3 binding when E2a is depleted (red box in B), but not other subcategories of Smad2/3 binding. D) Genes that retain Smad2/3 binding at the same genomic coordinates in E2a-depleted embryos (red box in B) are more likely to be downregulated by 2 fold or more in E2a-depleted embryos. E–F) Significant overlap exists between genes that are Smad2/3 targets and genes that are downregulated in E2a depleted embryos. Smad2/3 targets in which binding is at the same genomic position in control and E2a-depleted embryos are more likely to be downregulated in E2a-depleted embryos. These genes maintain enrichment for DAVID terms associated with early development. See also Tables S1, S2.
