Figure 1.
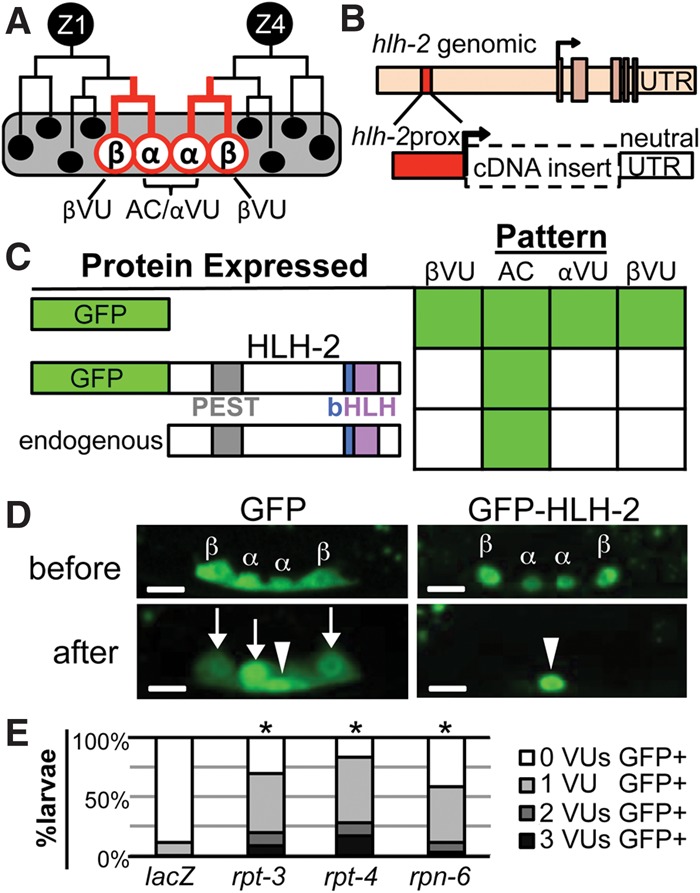
HLH-2 is post-translationally degraded in VUs. (A) Two cells, Z1 and Z4, generate the cells of the somatic gonad primordium (Kimble and Hirsh 1979). The proximal region contains two α cells (Z1.ppp and Z4.aaa) and their sisters, the β cells. lin-12/Notch-mediated signaling between the α cells specifies one as the AC and the other as a VU. The β cells always become VUs; the βVU fate does not require lin-12 activity and is specified earlier than the αVU fate. (B) hlh-2prox (red box) is a 327-base-pair (bp) element that drives transcription of GFP exclusively in α and β cells and their parents (red lines in A) with a “neutral” unc-54 untranslated region (UTR). hlh-2prox continues to drive transcription after the AC and VUs have been specified, although, after primordium formation occurs, the αVU is brighter than the βVUs, which were specified earlier. (C) hlh-2prox::GFP-HLH-2 displays the same pattern of accumulation as Karp and Greenwald (2003) observed for endogenous HLH-2, indicating post-translational regulation of HLH-2 stability. As described in the Supplemental Material, we set a 400-msec exposure time as a stringent threshold for analysis to facilitate scoring large numbers of individuals; two to four independent transgenic lines were scored per construct (see Supplemental Fig. S9 for details). To present a large amount of data for individual animals scored in summary form, in all figures, solid green indicates that ≥80% of cells are GFP+, green stripes indicate that 20%–80% of cells are GFP+, and solid white indicates that ≤20% of cells are GFP+. (D) Representative photomicrographs of hlh-2prox::GFP (left) and hlh-2prox::GFP-HLH-2 (right) before (top) and after (bottom) primordium formation (see the Supplemental Material for staging details). After primordium formation, GFP-HLH-2 has been reliably turned over in all three VUs, but GFP alone is still readily detectable in all three VUs. In all photomicrographs, maximum intensity projection allows visualization of all four cells; an arrowhead marks the AC, and arrows mark the VUs. Bar, 5 μm. (E) RNAi targeting proteasome components rpt-3, rpt-4, and rpn-6 stabilizes GFP-HLH-2 in VUs (see the Supplemental Material for details). (*) P < 0.005 by two-tailed Fisher's exact test compared with lacZ(RNAi).
