Figure 1.
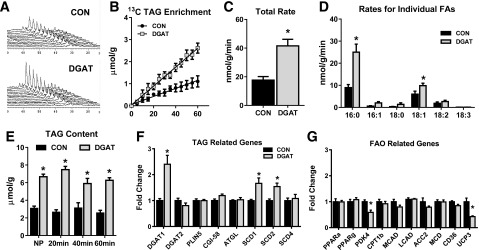
The effect of DGAT1 overexpression on cardiac TAG synthesis, content, and metabolic gene expression. A: Representative spectra from dynamic 13C NMR spectroscopy in isolated hearts perfused with a mixture of 13C LCFAs. B: Enrichment of 13C fatty acids into the TAG pool in lipid extracts from isolated perfused hearts (n = 3–4). C: Total incorporation rate of LCFAs into the TAG pool in isolated perfused hearts (n = 4–6). D: Incorporation rates for individual fatty acids (FAs) into the TAG pool. Rates for palmitate (16:0), palmitoleate (16:1), stearate (18:0), oleate (18:1), linoleate (18:2), and linolenate (18:3) are shown (n = 4–6 for each). E: TAG content measured in lipid extracts from nonperfused hearts (NP) and in isolated hearts perfused for 20, 40, or 60 min. Content is normalized to frozen heart tissue weight (n = 4–5). F and G: Expression of genes related to the TAG pathway and lipid metabolism in CON and MHC-DGAT1 (DGAT) hearts assessed via RT-PCR. *P < 0.05 vs. CON.
