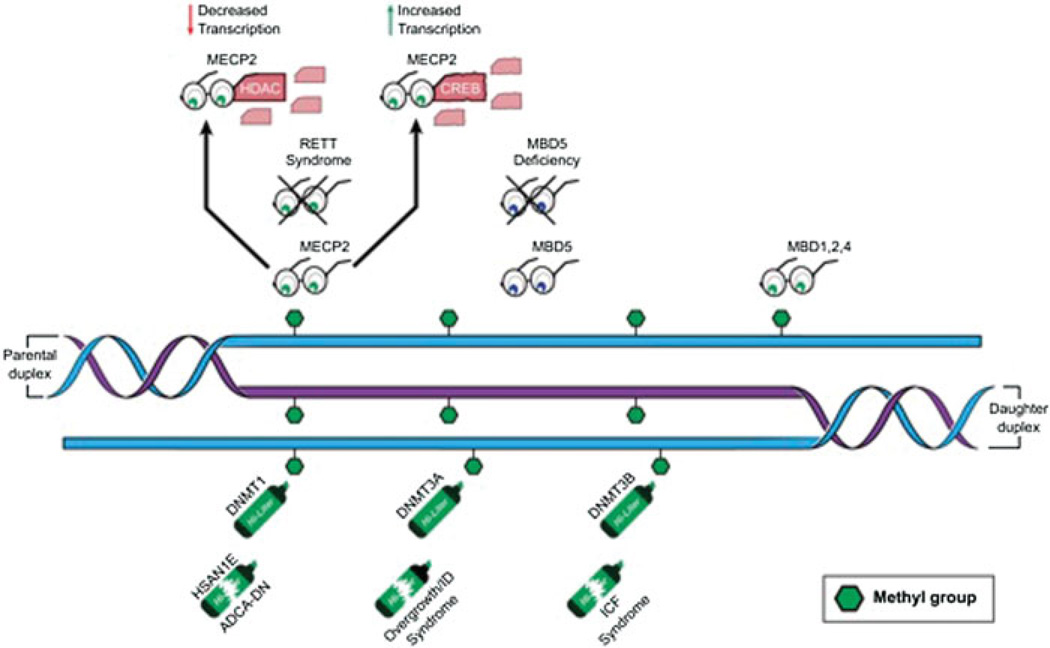
Fig. 1.
The DNA methylation machinery consists of DNA methyltransferases (highlighters), readers of the methylation mark (glasses), and erasers (not shown). Broken highlighters indicate diseases known to be caused by dysfunction of these proteins. Crossed out glasses indicate diseases caused by dysfunction of these proteins. DNMT1—DNA methyltransferase 1, is the maintenance methyltransferase that copies the signal from the parental strand; DNMT3A—DNA methyltransferase 3A and DNMT3B—DNA methyltransferase 3B are the de novo methyltransferases and are not limited to hemimethylated sites; MECP2—methyl-CPG binding protein reads the DNA methylation mark and can either lead to gene activation or repression depending on partners; MBD1, 2, 4—methyl-binding proteins 1, 2, and 4 also read methyl-CpGs; MBD5—methyl-binding protein 5 does not read a CpG methylation, but associates with heterochromatin; HSAN1E—hereditary sensory and autonomic neuropathy type 1 with dementia and hearing loss syndrome; ADCA-DN—autosomal dominant cerebellar ataxia, deafness, and narcolepsy syndrome; ICF—immunodeficiency, centromeric instability, and facial anomalies syndrome; HDAC—histone deacetylase protein; CREB—cAMP-binding response element-binding protein.