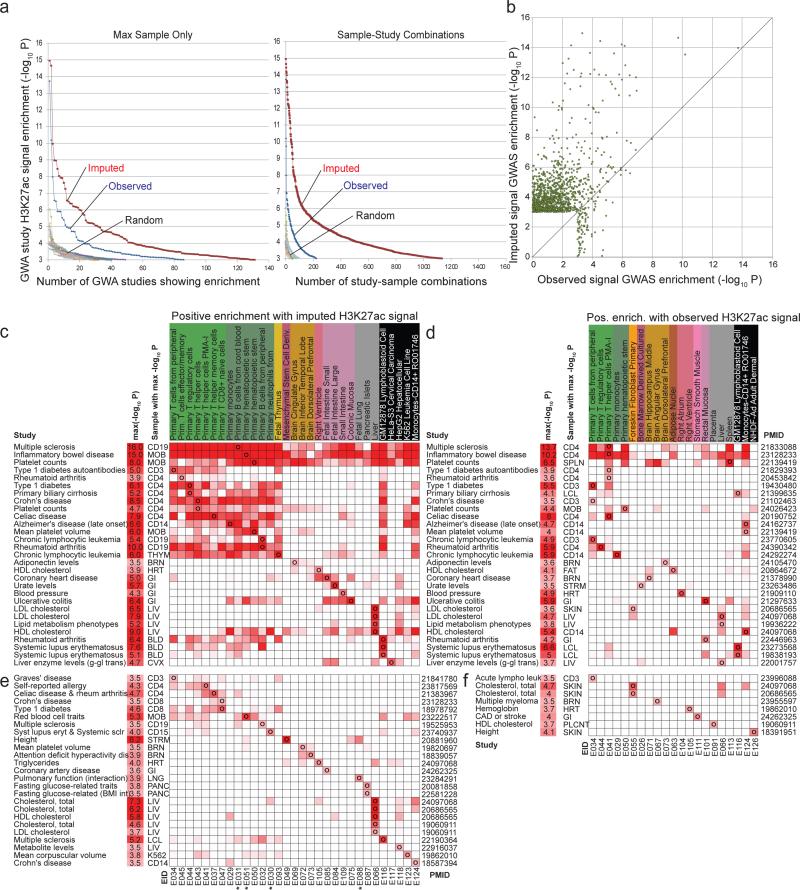
Figure 4. Overlap with trait-associated genetic variants from GWAS.
(a) (Left) The x-axis shows the number of genome-wide association studies for which there was at least one sample for which the H3K27ac signal was significantly different than based on a background of all GWAS catalog SNPs at significance level indicated on the y-axis using a Mann-whitney U Test (see Methods). This is shown for the observed data (blue), the imputed data restricted to the 98 samples with observed data (red), and the observed and imputed data based on ten randomizations of the GWAS catalog. (Right) The same as on left, but counting study-sample combinations opposed to just studies. (b) A scatter plot showing the –log10 p-value computed for each study-sample combination based on the observed data (x-axis) and imputed data (y-axis) for each combination that had a p-value of 10−3 or better based on either the imputed or the observed data for H3K27ac. The diagonal line is the y=x line showing most of the highest significant studies based on either the observed or imputed data are above it. Additional marks can be found in Fig. S24-26. (c-f) Enrichment matrices (heatmaps) showing all studies (rows) with uncorrected -log10 p-value ≥3.5 for at least one reference epigenome (columns) based on H3K27ac imputed data c,e and observed data d,f. For each study (rows) is shown the trait, most-significant p-value (-log10 p), max-sample abbreviation (Abbr), and pubmed identifier (PMID). Only samples that showed the highest-significance positive enrichment for at least one study are shown. c,d: studies that were significant (-logP≥3.5) for both observed and imputed. Top three rows show studies with broad enrichment across samples. e,f: Same enrichments for studies that were only significantly enriched using imputed (e) or observed (f) H3K27ac signal. Stars denote H3K27ac signal tracks that only exist as imputed data. Expanded enrichments for all samples, all Tier-1 marks, and additional GWA studies are in Table S2.