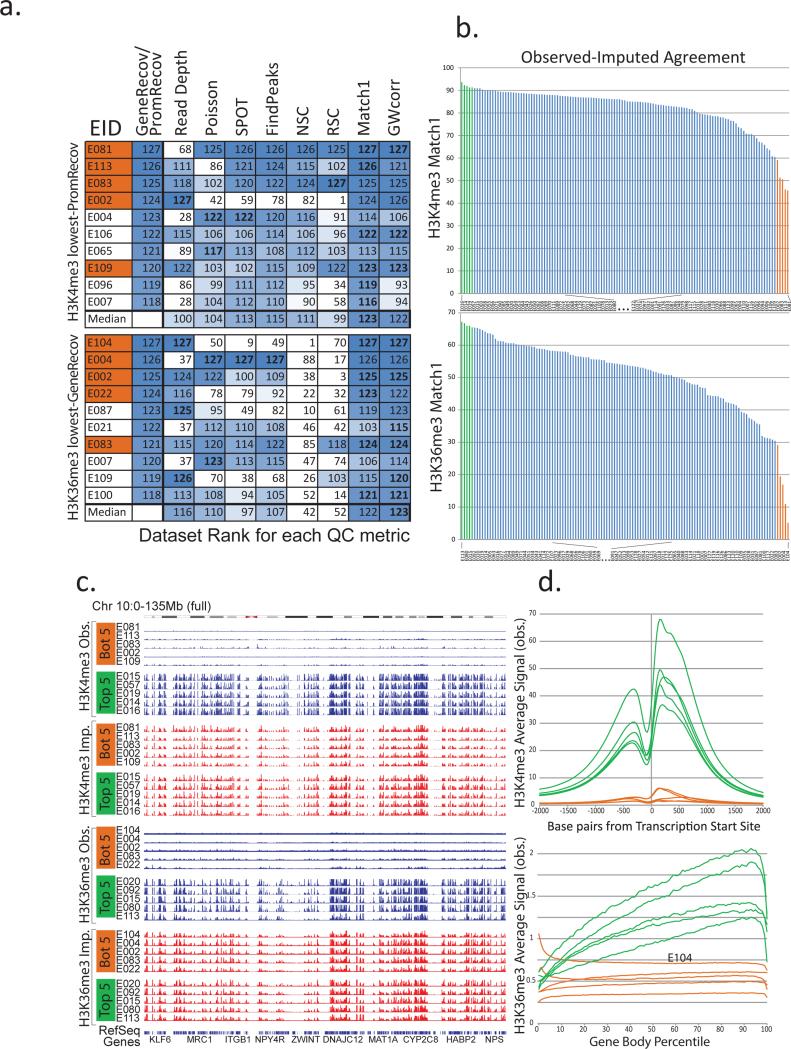
Figure 5. Low similarity between imputed and observed data reveals low-quality datasets.
(a) Comparison of quality control (QC) metrics (columns) for the ten datasets (rows) showing lowest agreement with gene and TSS annotations (Fig. 3a,b), based on H3K4me3 PromRecov (top) and H3K36me3 GeneRecov (bottom). Each entry shows rank (out of 127) for PromRecov/GeneRecov, read depth, and each QC metric (Poisson statistic, Signal Proportion of Tags, FindPeaks, Normalized and Relative Strand Correlation between forward and reverse strands (NSC and RSC)), and similarity between imputed and observed data (Match1 and GWcorr). Orange-shaded EIDs denote the five worst-agreement datasets from panel b. Data sets with the same read depth, (due to highly sequenced data sets being previously downsampled to the same number of reads10), are given the same expected rank if ties were broken randomly. Most-problematic datasets (based on lack of gene or +/−2kb TSS annotation recovery) are sometimes missed by traditional QC measures, but consistently show low imputation agreement. (b) Distribution of agreement between top 1% observed signal and top 1% imputed signal locations for H3K4me3 (top) and H3K36me3 (bottom), highlighting five worst-similarity (orange) and five highest-similarity (green) datasets. (c) Observed (blue) and imputed (red) signal tracks for worst-similarity (orange) and best-similarity (green) datasets for H3K4me3 (top) and H3K36me3 (bottom) for the entire chromosome 10 (0-135Mb). Datasets with the lowest agreement have relatively flat signal, suggesting that when observed and imputed datasets disagree most, it is usually the observed datasets that are of lowest quality. (d) Aggregation of observed signal for H3K4me3 in TSS (top) and H3K36me3 in gene bodies (bottom) for the 5 best-agreement (green) and worst-agreement (orange) datasets, highlighting the unusual profiles of some worst-agreement datasets, suggesting they are of lower quality, even though they were not flagged by traditional QC metrics.