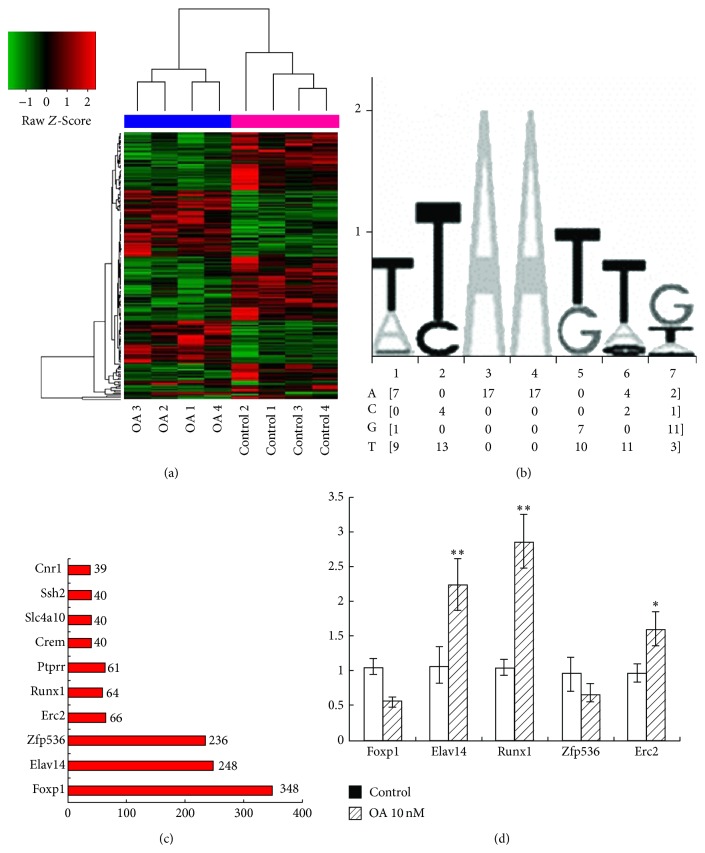
Figure 4.
Genes differentially regulated by OA and bioinformatical analysis of these genes. Single NSCs at density of 5000 cells/well in 6-well plate were cultured in differentiation medium. Cells were exposed to 10 nM OA or not. 48 h later, cells were harvested. The total mRNA was extracted followed by whole genome mRNA expression measurement. 183 genes were differentially regulated by OA. Among 183 genes, 87 were predicted under the control of Nkx-2.5. Using these 87 genes, hierarchical clustering analysis was performed (a). The upstream regulation sequences among these 87 genes shared common predicted motif for Nkx-2.5 binding. The frequency matrix for this motif was shown. TTAATTG was the most frequent pattern (b). The number of binding sites existing in the upstream regulatory sequence of 87 genes was calculated. Top 10 genes with the largest numbers were shown (c). The expression of 5 differentially regulated genes including Foxp1, Elavl4, Runx1, Znf536, and Erc2 was confirmed by quantitative real-time PCR (d).