Fig. 3.
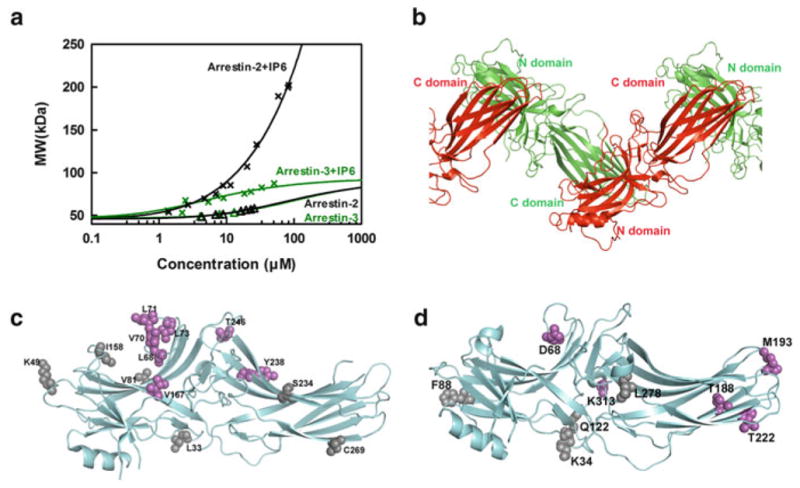
The two nonvisual arrestins form distinct oligomers. (a) The average molecular weight of arrestin-2 and arrestin-3 in the presence (crosses) and absence (triangles) of 100 μM IP6 as a function of total arrestin concentration was measured by MALLS. Arrestin-2 data were fit by a linear polymerization model (black line), while arrestin-3 data were fit by a monomer–dimer model (green line). Neither nonvisual arrestin showed a propensity to self-associate at physiologically relevant concentrations in the absence of IP6. (b) The crystal structure of arrestin-2 in complex with IP6 [PDB ID: 1ZSH (Milano et al. 2006)] shows that arrestin-2 forms “infinite” chains through C-to-N-domain interactions mediated by IP6. (c) The positions of spin-labeled sites are shown as spheres on the crystal structure of arrestin-2 [PDB ID: 1ZSH (Milano et al. 2006)]. The sites with inter-subunit distances shorter than 50 Å (Leu68, Val70, Leu71, Leu73, Val167, Tyr238, and Thr246), as measured by DEER spectroscopy in the presence of IP6, are colored magenta, and the ones with inter-subunit distance longer than 50 Å (Leu33, Lys49, Val81, Ile158, Ser234, and Cys269) are colored gray [data from Chen et al. (2013)]. (d) The positions of spin-labeled sites are shown as spheres on the crystal structure of arrestin-3 [PDBID: 3P2D (Zhan et al. 2011)]. The sites with inter-subunit distance shorter than 50 Å (Asp68, Thr188, Met193, Thr222, and Lys313), as measured by DEER spectroscopy in the presence of IP6, are colored magenta, while the ones with inter-subunit distance longer than 50 Å (Lys34, Phe88, Gln122, and Leu278) are colored gray [data from Chen et al. (2013)]
