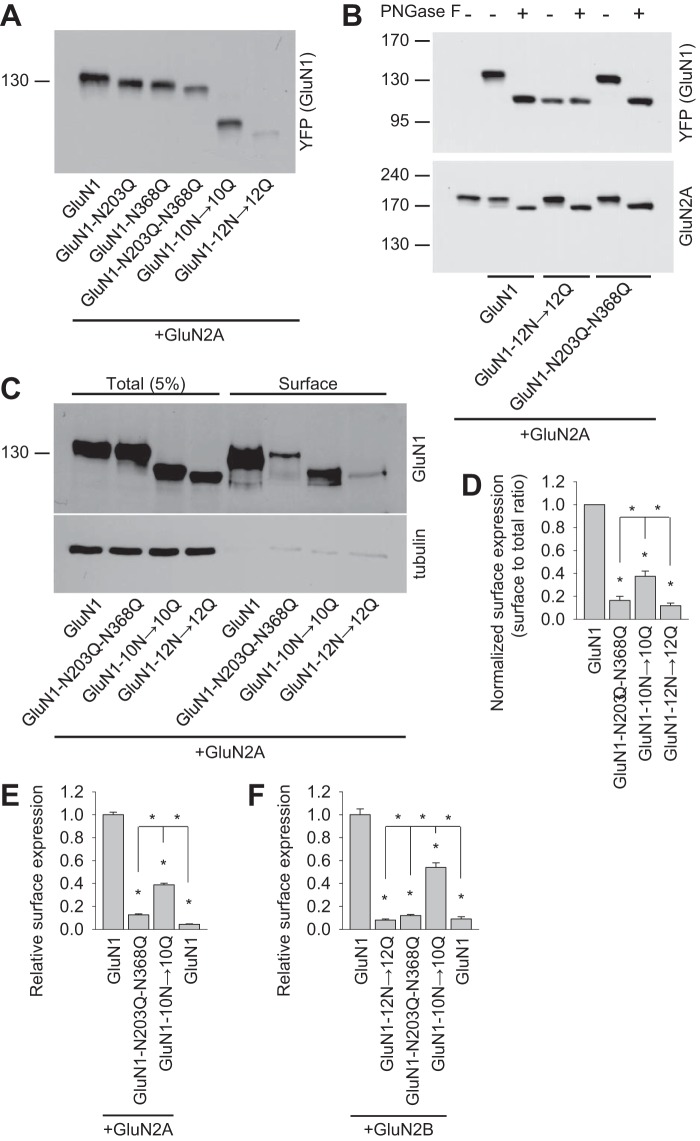
FIGURE 2.
Biochemical characterization of NMDARs containing GluN1 subunits that lack specific N-glycosylation sites. A, HEK293 cells were cotransfected with either wild-type or mutant YFP-GluN1-1a subunits (GluN1) together with the GluN2A subunit. 24 h after transfection, the GluN1 subunit was examined by Western blot analysis. B, HEK293 cells were cotransfected with either wild-type or mutant YFP-GluN1-1a subunits (GluN1) together with the GluN2A subunit. 24 h after transfection, cell homogenates were incubated in the presence or absence of PNGase F and analyzed by Western blot using either the anti-GFP antibody (to detect GluN1) or the anti-GluN2A antibody. C, cell surface biotinylation assay for the indicated YFP-GluN1-1a (GluN1) subunits coexpressed with the GluN2A subunit. HEK293 cells were transfected with the indicated YFP-GluN1-1a and GluN2A subunits. The surface receptors were biotinylated and pulled down using streptavidin-agarose resin. Total input (5% of the lysate) and the surface GluN1 receptor were detected using anti-GluN1 antibody. Anti-tubulin antibody was used to confirm the integrity of the assay. D, summary of the experiments in C. The band intensities of the surface and total NMDAR pools were quantified as described under “Experimental Procedures” (n = 5). *, p < 0.05 versus wild-type GluN1/GluN2A, analyzed by one-way ANOVA. E and F, COS-7 cells expressing the indicated wild-type or mutant YFP-GluN1-1a subunit (GluN1) with or without the GluN2A (E) or GluN2B (F) subunits were labeled using anti-GFP under non-permeabilizing and permeabilizing conditions. Expression was measured using a quantitative colorimetric assay. The bar graphs show the relative surface expression of the indicated GluN subunit combinations from three independent experiments (n = 9). *, p < 0.05 versus wild-type GluN1/GluN2A or GluN1/GluN2B receptors, analyzed by one-way ANOVA.