FIGURE 6.
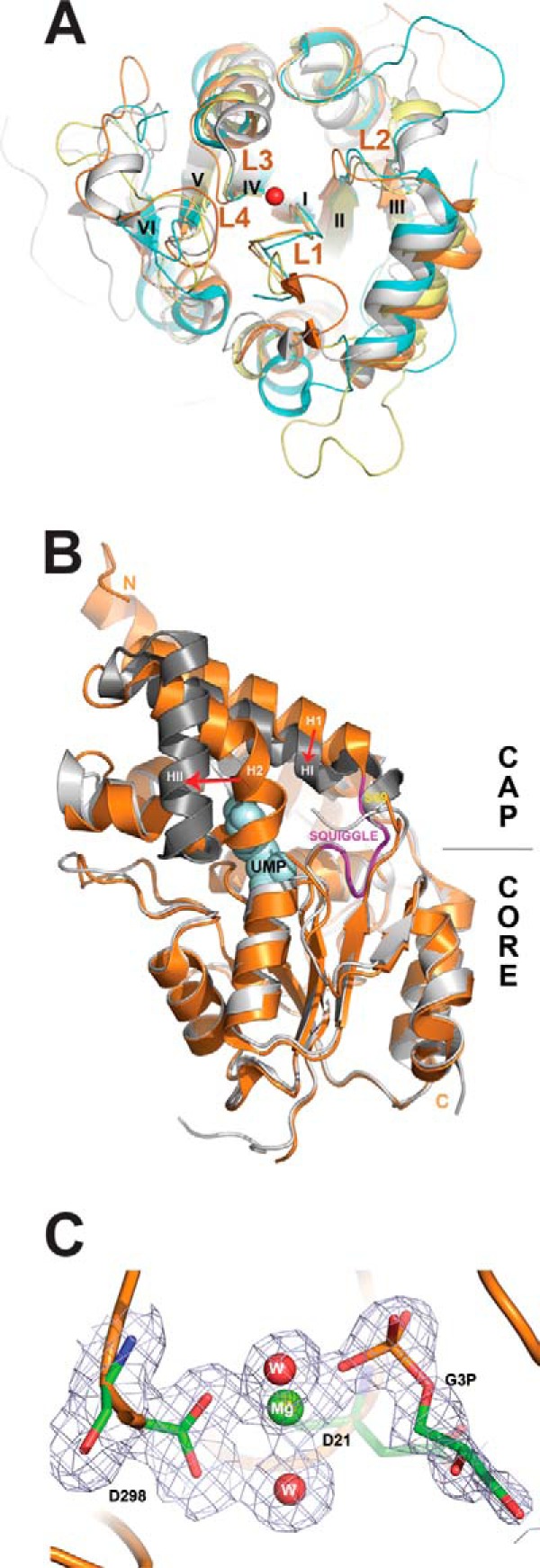
Structural analysis of S. cerevisiae HADs. A, structural superposition of core domains of the four yeast HADs: RHR2 (gray), SDT1 (cyan), UTR4 (magenta), and YKR070W (orange). The overlay shows close alignment of the structural elements involved in catalysis, including the active site loops (L1–L4) and bound metal ions (the Mg2+ ion from the YKR070W structure is shown as a red sphere). The conserved structural elements squiggle and flap are positioned close to loop L1 (labeled). For clarity, the cap domains of these HADs are not shown, whereas the β-strands are numbered with Roman numerals. B, structural superposition of the apo-form and UMP binary complex of SDT1. We have determined the 1.70 Å structure of the apo-form of SDT1 (PDB 3NUQ; orange ribbon), whereas the structure of the SDT1-substrate complex with UMP is available from PDB (3OPX; shown as a gray ribbon with UMP shown as a space-filled model). Superimposition of these two structures revealed a similar overall structural fold (r.m.s. deviation 1.43 Å) with significant structural changes in the cap domain (indicated by red arrows). C, close-up view of the active site of YKR070W with bound Gly-3-P with 2Fo − Fc electron density map (gray) contoured at 1.0 σ. The bound Gly-3-P and residues are shown as sticks, the Mg2+ ion as a green sphere, and two water molecules as red spheres. The molecule of Gly-3-P was omitted during the map calculation.
