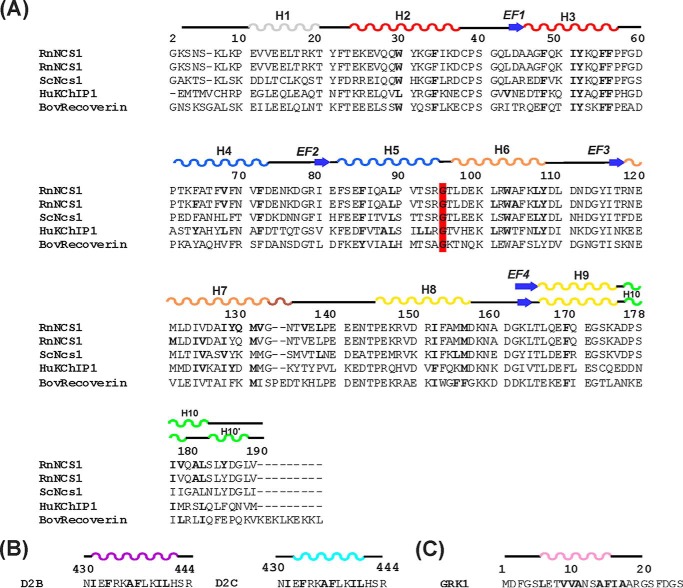
FIGURE 2.
Summary of interaction sites of NCS proteins. A, alignment of the primary sequence of rat NCS-1 (RnNCS-1; Swiss-Prot P62168) with Saccharomyces cerevisiae Ncs1 (ScNcs1; Swiss-Prot Q06389), human KChIP1 (HuKChIP1; Swiss-Prot Q9NZI2.2) and bovine recoverin (BovRecoverin; Swiss-Prot P21547). RnNCS-1 is repeated to separately identify NCS-1 residues that form intermolecular interactions with D2R (top sequence) and GRK1 (2nd sequence) peptides. Numbering of amino acids is in accordance to the primary sequence of rat NCS-1. Secondary structure elements are derived from the structures of rat NCS-1 in complex with D2 receptor peptide (PDB 5AER). The C-terminal region forms helix 10 in the NCS-1 apoprotein and in the NCS-1·GRK1 complex (denoted by the upper line of secondary structure for this region). In the D2R peptide complex, helix 10 is replaced by a helix-turn-helix, and this is denoted as helices 10 and 10′. The four EF-hands, EF1, EF2, EF3, and EF4, are colored red, blue, orange and yellow, respectively. The short 310 helix between EF3 and EF4 is colored brown and the C-terminal region in green. The hydrophobic residues that form interactions in the different complexes are highlighted in boldface; the different complexes are NCS-1·D2R complex (PDB code 5AER), the RnNCS-1·GRK1 complex, ScNcs1·Pik1 (PDB Code 2JU0), HuKChIP1·Kv4.3 (PDB Codes 2NZ0 and 2I2R) and BovRecoverin/rhodopsin kinase (PDB code 2I94). The conserved Gly-95 that divides NCS proteins into the N- and C-lobes is highlighted in red. The hydrophobic contacts are analyzed from the deposited structures using the Protein Interactions Calculator Webserver (38). B and C, amino acid sequence of D2 dopamine receptor (B) and GRK1 (C) peptides with residues involved in hydrophobic interactions with NCS-1 in boldface and the α-helices colored in magenta/cyan and pink, respectively, in the D2R and GRK1 complex. In the NCS-1·GRK1 peptide structure, a hydrophobic triad is formed between Met-156, Ile-179, and the peptide residue Ile-16.