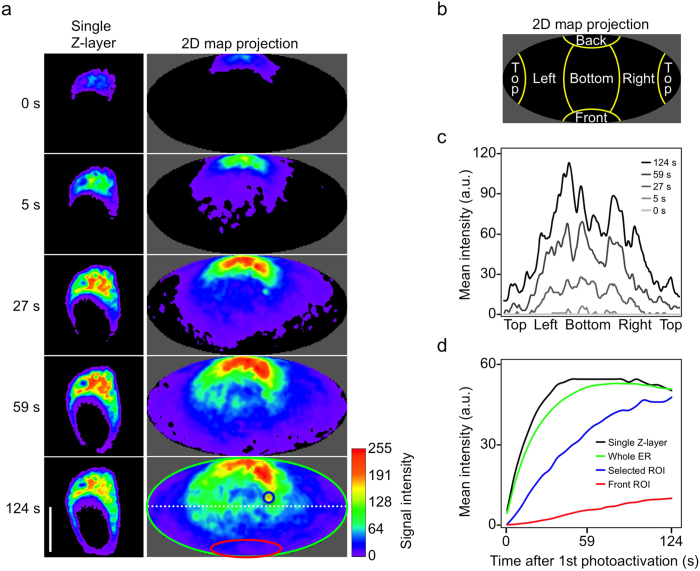
Figure 3. Quantitative analyses with map projections.
(a) Photoactivation experiment of an N2a cell expressing CD3δ-PAmCherry, displayed as a single Z-layer (left) or Mollweide map projection (right). Different time points upon continuous photoactivation are shown, starting at 0 s immediately after the first photoactivation event at the back-lateral side of the cell. The (white dotted) line and (red, green and blue) area selections used for fluorescence signal plot profiling (c) and mean intensity area measurements (d) are displayed on the map projection at time point 124 s. For orientation, a schematic Mollweide projection (b) shows the position of the ER signal with respect to the nucleus as four lateral sides (left, right, back, front) and top and bottom positions (above and below the nucleus). (c) Quantitative dynamic analysis of photoactivated CD3δ-PAmCherry diffusion across the full mid-horizontal section of the ER by plot profiling on the map projections shown in (a). (d) Quantitative analysis of photoactivated CD3δ-PAmCherry at different ROIs over time. The colors of the mean intensity curves correspond to the colors used for the selections shown in (a). All mean intensities were measured on the original image stacks by cross-referencing the area information from the corresponding maps. For the single Z-layer images, the whole ER area from that single layer was used for measurements. The Mollweide maps are presented as pixel-interpolated, maximum intensity projections. For better intensity discrimination, all images in (a) are displayed by using the rainbow smooth lookup table. a.u., arbitrary units. Scale bar, 10 μm.