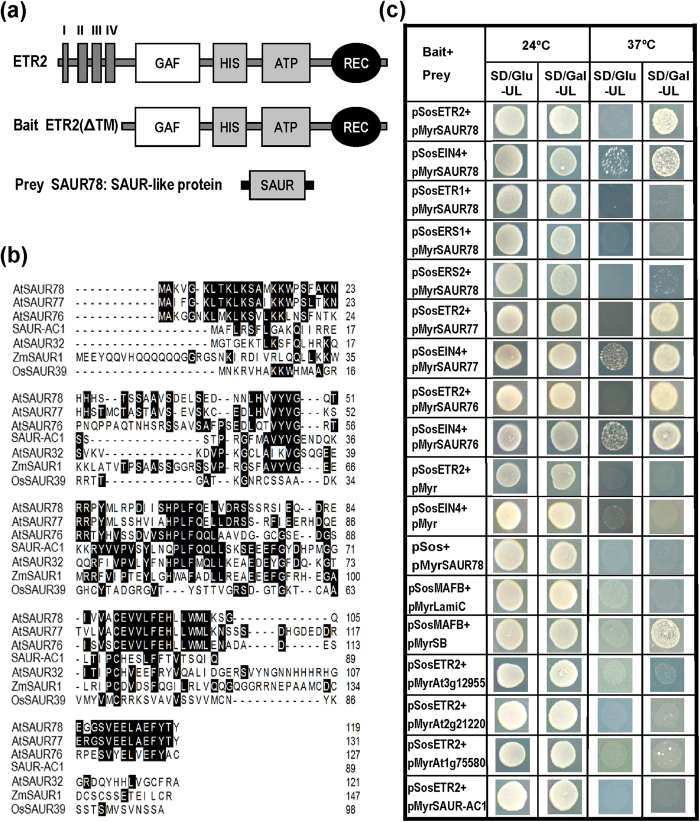
Figure 1. Identification of ethylene receptor-interacting proteins SAURs in Arabidopsis.
(a) Schematic representation of the bait ETR2 and the prey identified. I, II, III and IV indicate putative transmembrane regions. GAF: conserved domain originally found in cGMP-binding phosphodiesterases, cyanobacterial adenylyl cyclases, and a formate-hydrogen lyase transcription activator from E. coli; HIS: H-containing domain; ATP: ATP-binding domain; HIS plus ATP constitute the kinase domain; REC: receiver domain. The region without transmembrane segments was used as bait to screen cDNA library in yeast two-hybrid assay. SAUR78 was identified as an ETR2-interacting protein. (b)Alignment of SAUR78 as well as its close homologues SAUR76 and SAUR77 with other known SAUR proteins. SAUR-AC1 and AtSAUR32 are from Arabidopsis. ZmSAUR1 is from maize and OsSAUR39 is from rice. Amino acids shaded in black indicate identity. (c) Interactions of the three SAURs with Arabidopsis ethylene receptors ETR2 and EIN4 in yeast two-hybrid assay. The four other SAUR proteins At3g12955, At2g21220, At1g75580 and SAUR-AC1(At4g38850), which are not grouped with the three SAURs, did not show positive interactions with ETR2. At 24 °C, all the yeast transformants can grow. At 37 °C, growth of transformants on SD/Gal-UL but not on SD/Glu-UL indicates positive interaction. The pSosMAFB plus pMyrSB indicate positive interaction control while pSosMAFB plus pMyrLamiC and other combinations with pMyr or pSos vectors served as negative controls.