Figure 4.
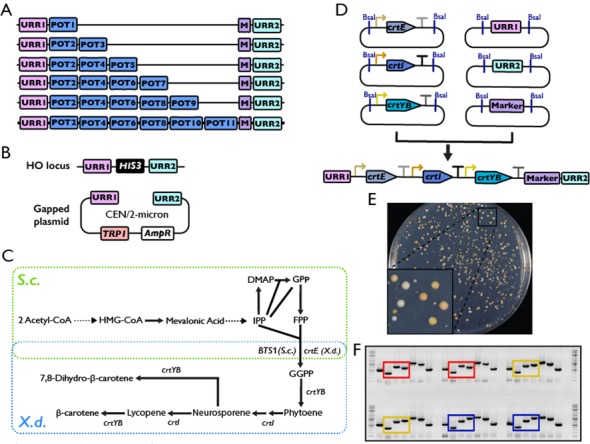
Hierarchical assembly of metabolic pathways. (A) Graphical representation of assembling a pathway with up to six genes by cherry-picking the eleven POT vectors. Different POT vectors should be used based on the number of genes to be assembled. (B) Different targets designed to accommodate the exogenous pathways. One is at the HO locus on chromosome IV, where a selectable marker (HIS3 as shown) flanked by the URRs is engineered. The other is an ectopic plasmid, either at low copy (CEN plasmid/pRS414-URR) or at high copy (the 2-micron plasmid/pRS424-URR). (C) Overall scheme for engineering β-carotene production in S. cerevisiae. Genes from X. dendrorhous are circled in blue. IPP, isopentenyl diphosphate; DMAP, dimethylallyl diphosphate; GPP, geranyl diphosphate; FPP, farnesyl diphosphate; GGPP, geranylgeranyl diphosphate. (D) Hierarchical assembly of β-carotene biosynthesis pathway. The assembled TUs (from Figure 3) in the POT vectors and URR1/URR2/Marker were digested with restriction enzymes, and ligated in vitro to the DNA fragment containing the complete pathway, which was used to transform the yeast directly. (E) The S. cerevisiae strain growing on the selective plate after transformation. The presence of yellow colonies indicates the cells could produce β-carotene successfully. The plate was photographed after 60 h of incubation at 30°C. (F) The diagnostic PCR of the S. cerevisiae transformants. Those circled in red are the cells containing URR1-pCYC1-crtE-tADH1-pTEF2-crtI-tADH1-pCYC1-crtYB-tADH1; those circled in yellow are the cells containing URR1-pCYC1-crtE-tADH1-pTEF2-crtItADH1-pTEF2-crtYB-tADH1; and those circled in blue are the cells containing URR1-pCYC1-crtE-tADH1-pTEF2-crtI-tADH1-pTDH3-crtYB-tADH1.
