Figure 1.
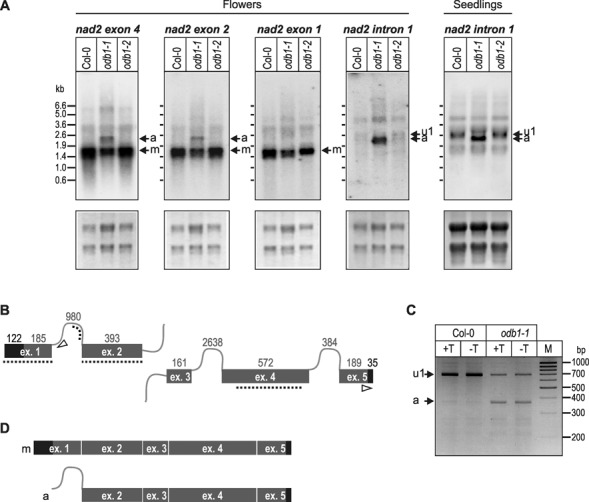
Incompletely spliced nad2 transcripts over-accumulate in Arabidopsis plants lacking ODB1. (A) Probes for exon 4, exon 2, exon 1 and intron 1 of the mitochondrial nad2 gene were hybridised to filter-immobilised total RNA isolated from odb1–1, odb1–2 and wild-type (Col-0) flowers and seedlings (top panels). RNA marker sizes are indicated. Signals corresponding to the mature nad2 mRNA (m), an alternative transcript of about 2300 nt over-accumulating in odb1–1 (a) and transcripts from which the first intron has not been removed (u1) are indicated by arrows. The same filters stained with methylene blue are shown as loading controls in the bottom panels. (B) The nad2 gene has five exons that are transcribed from two mtDNA regions and joined by trans-splicing of intron 2. Lengths of exons and cis-spliced introns are given in nucleotides. Only exons are drawn to scale (grey, coding regions; black, untranslated regions); introns are depicted as thin grey lines. Dotted lines mark probe target regions. Arrowheads indicate positions of primers used in the cRT-PCR experiment shown in (C). (C) The 5′ end of the nad2 transcript over-accumulating in odb1–1 was identified by cRT-PCR performed on TAP-treated (+T) and untreated (−T) seedling RNA. Products obtained for odb1–1 and the wild type (Col-0) were separated on agarose gels alongside a molecular weight marker (M), cloned and sequenced. A product seen for both genotypes corresponded to nad2 transcripts retaining the first intron (u1). A smaller product preferentially amplified from odb1–1 (a) corresponded to a nad2 transcript retaining intron 1 but lacking exon 1. With a length of 2330 nt this transcript matches the size of the odb1–1-specific transcript seen in RNA gel blots in (A). Amplification of the products labelled a and u1 strictly depended on the RNA ligation step during the cRT-PCR procedure (Supplementary Figure S1). (D) Schematic illustrating the structure of the nad2 transcript over-accumulating in odb1–1 (a) compared with the mature nad2 mRNA (m).
