Figure 5.
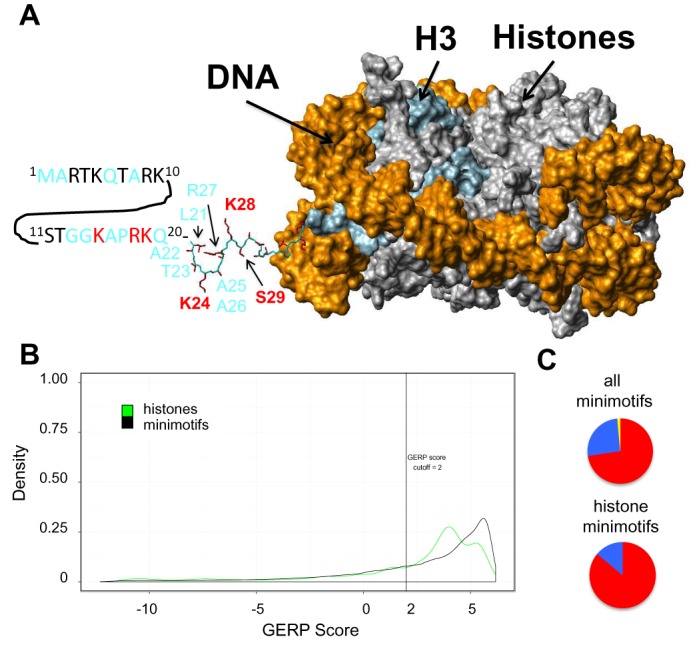
SNPs present in histone minimotifs. (A) Surface plot of nucleosome showing locations of polymorphic minimotif residues. Histone H3 is colored light blue and DNA is colored gold. The structure is from 1AOI and accession number for the sequence is P02302. A portion of the N terminal tail of histone H3 that resolved in the crystal structure is shown as a stick figure with side chains colored red. The sequence of the portion of the N-terminal tail that did not resolve (residues 1–20) is shown as a sequence using single letter amino acid code. Labels in black or red bold font indicate sites of covalent modification in minimotifs. Fonts colored black are conserved and those colored red are polymorphic. Fonts colored cyan are neither polymorphic nor post-translationally modified. No polymorphic residues were located inside the core of histone H3. (B) Density plots for GERP scores for minimotifs present in the 53 human histones. A vertical line indicates the GERP score threshold of 2. (C) Pie graphs showing the percentages of histone minimotif SNPs groups under neutral, negative and positive selection as measured by the SLR statistic in Figure 2.
