Figure 8.
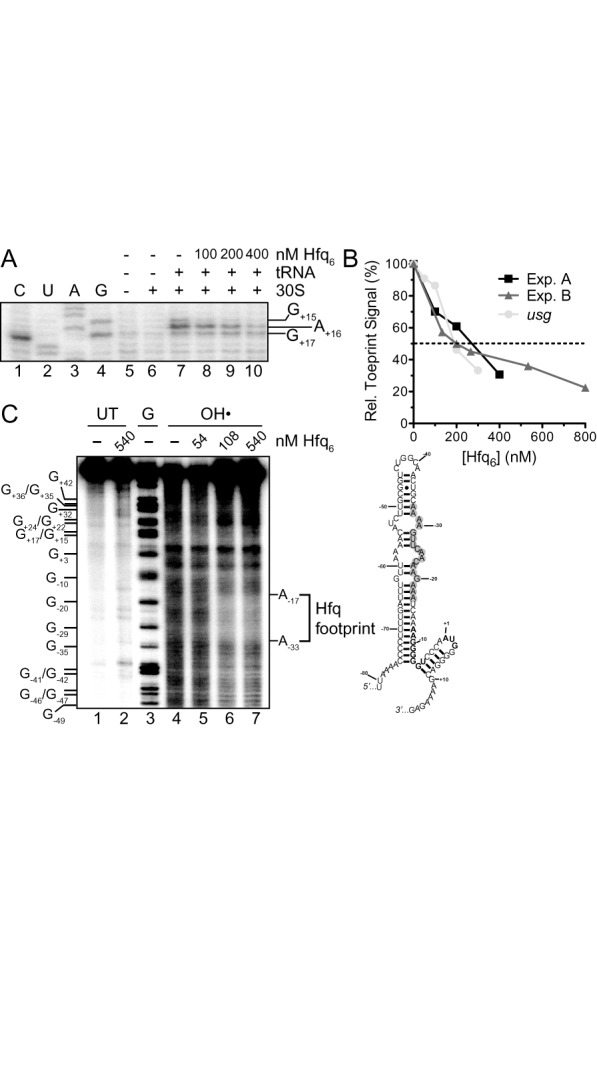
Hfq inhibits 30S ribosomal subunit binding to tnpA and binds upstream of the SD. (A) Toeprint assay showing the effect of Hfq on 30S ribosomal subunit binding to tnpA. Hfq (100–400 nM; hexamer concentration) was added to tnpA (200 nM) prior to addition of 30S ribosomal subunit and initiator tRNA. A section of the gel image including the toeprint signal is shown. (B) The percent inhibition of toeprint signal upon incubating Hfq with tnpA or an mRNA that does not interact with Hfq (usg, (48)) is shown; the usg data come from ref (40). For both mRNAs the toeprint signal in the absence of Hfq was set at 100%. Experiment A refers to part (A) of this figure while Experiment B refers to Supplementary Figure S4. (C) Hydroxyl radical footprinting experiment with 5'32P-labeled tnpA1–173 (68 nM) and the indicated concentrations of Hfq. Subsequent to mixing tnpA and Hfq limited RNA cleavage by hydroxyl radical treatment was carried out as previously described (7). UT is untreated RNA and G is an RNA cleavage ladder produced by RNase T1 cleavage. The Hfq footprint between residues −17 and −33 defines an Hfq binding site in tnpA and the position of this site is highlighted (gray circles) in our model for tnpA1–173; the tnpA SD and start codon are in bold. Note that a version of this gel image was previously published in Methods in Molecular Biology (38).
