Figure 1.
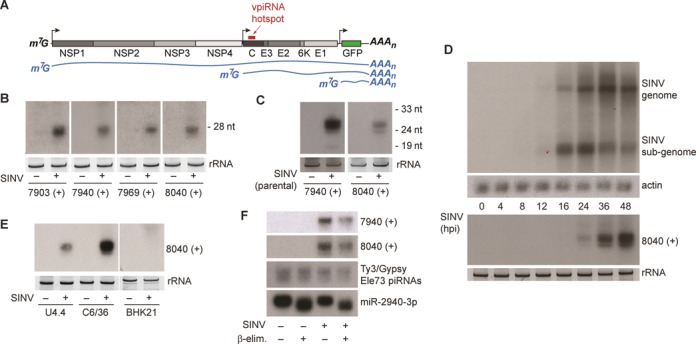
Selected mature vpiRNAs are abundant in Aag2 cells. (A) Schematic representation of the SINV-GFP genome. The individual viral proteins are indicated in gray and the position of the piRNA hotspot is marked by the red bar. The blue lines show the three (+) strand RNA species that can be found in infected cells. (B) Small RNA northern blot of four vpiRNAs in uninfected or SINV infected Aag2 cells. Probe names indicate the 5′ end position of the detected vpiRNAs, which are all derived from the SINV (+) strand. (C) Small RNA northern blot for vpiRNA in uninfected or SINV (parental virus) infected Aag2 cells. (D) Northern blot analysis of viral genomic and subgenomic RNA (upper panel) or vpiRNAs (lower panel) using a probe against vpiRNA 8040 (+). Probing for actin mRNA serves as loading control. (E) Northern blot analysis of vpiRNA in uninfected or SINV-infected Aedes albopictus mosquito cells (U4.4 and C6/36) and baby-hamster kidney cells (BHK21). For small RNA northern blots in panels B to E, ethidium bromide staining of ribosomal RNA (rRNA) serves as loading control. In panel B the loading controls for 7903 (+) and 7940 (+) are identical, since the same membrane was subsequently hybridized to these probes after harsh stripping in hot 0.1% SDS. (F) Northern blot detection of vpiRNAs, Ty3/Gypsy element 73 transposon piRNAs or miR2940–3p. Before blotting, β-elimination was performed on total RNA as indicated.
