Figure 3.
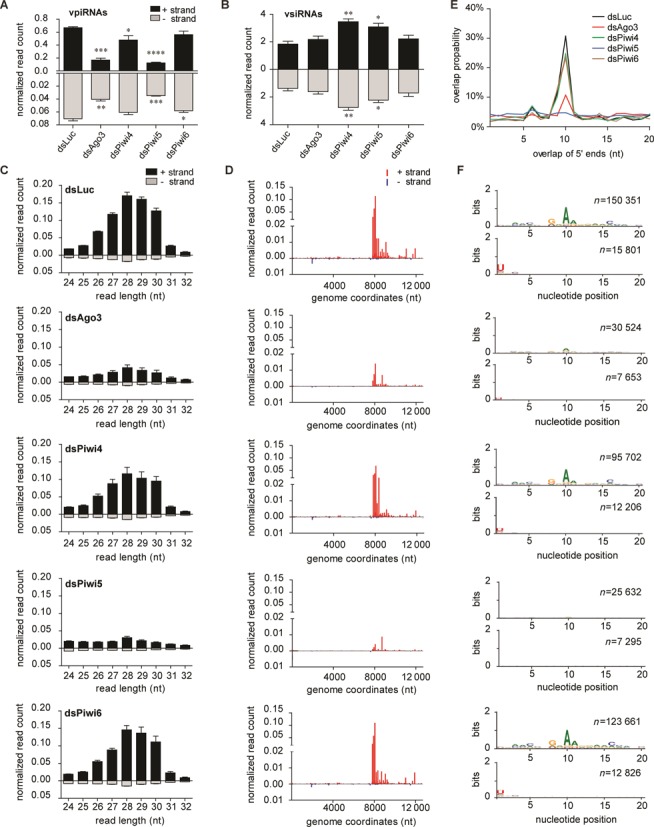
Piwi5 and Ago3 are required for vpiRNA biogenesis. (A, B) Number of 25–30 nt piRNA reads (A) and 21-nt siRNA reads (B) derived from the SINV (+) strand (black bars) and (−) strand (gray bars) in the indicated PIWI-protein knockdown libraries. Two-tailed student's t-test was used to determine statistical significance (*P < 0.05; **P < 0.01; ***P < 0.001: ****P < 0.0001). (C) Size profile of small RNAs mapping to the (+) strand (black bars) or the (−) strand (gray bars) of SINV. Bars in A–C are the mean +/− SEM of the three independent libraries. (D) Genome distribution of 25–30 nt small RNAs across the (+) strand (red) or (−) strand (blue) of the SINV genome. The average counts (three experiments) of the 5′ ends of the small RNA reads at each nucleotide position are shown. (E) The mean probability (n = 3) for 5′ overlaps between viral piRNAs from opposite strands in the indicated knockdown libraries. (F) Nucleotide bias at each position in the 25–30 nt small RNA reads mapping to the SINV (+) strand (upper panels) and (−) strand (lower panels). All reads of three independent experiments were combined to generate the sequence logo; n, number of reads.
