Figure 4.
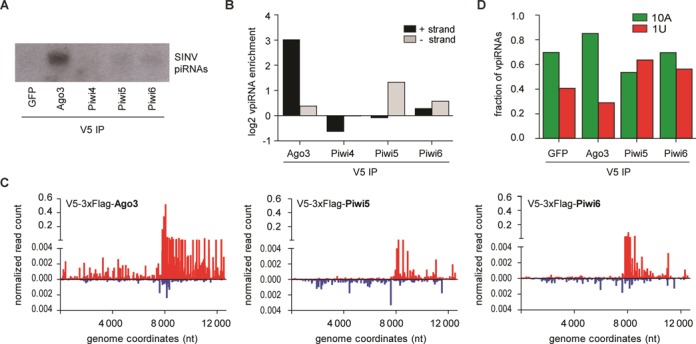
Association of vpiRNAs with individual PIWI proteins. (A) Northern blot analysis of vpiRNAs in RNA isolated from IPs of the indicated V5-epitope tagged proteins. Viral piRNAs were detected using a pool of the four probes presented in Figure 1A. (B) Enrichment of 25–30 nt small RNAs from the SINV (+) strand (black bars) or (−) strand (gray bars) in the IP of the indicated V5-epitope tagged PIWI proteins compared to the V5-tagged GFP-IP. (C) Distribution of 25–30 nt small RNAs in the indicated PIWI IPs across the (+) strand (red) or (−) strand (blue) of the SINV genome. Every data point shows the number of reads at each nucleotide position normalized against the size of the library (% of library). To account for background binding, the normalized read counts of the GFP-IP at each position were subtracted. (D) Fraction of 25–30 nt SINV-derived small RNA reads from the indicated deep-sequencing libraries that have an adenine at position 10 (10A; green bars) or uridine at position 1 (1U; red bars), respectively. No data for Piwi4 is shown in panels C and D since the V5-IP for this protein was not enriched for vpiRNAs.
