Figure 2.
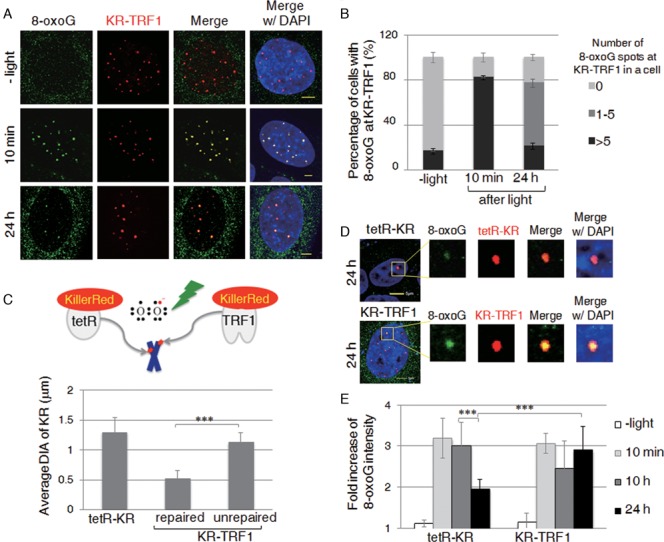
Long telomeric repeats have less efficient DNA repair compared to chromosomal DNA with a similar amount of DNA damage. (A) Staining of 8-oxoG in KR-TRF1-expressing U2OS cells with or without light activation. Cells were collected after recovery in the dark for the indicated times. Scale bar equals 5 μm. (B) Percentage of cells showing co-localization of 8-oxoG with KR-TRF1 spots, respectively. Each data point was derived from >150 cells in three independent experiments. Error bars depict standard deviations. (C) Schematic for generating single genome locus damage using tetR-KR and using KR-TRF1 to target damage to telomeres. Quantification of average diameters (DIA) of tetR-KR and KR-TRF1 spots with or without co-localization with 8-oxoG in KR-TRF1-expressing U2OS cells after 24 h recovery in the dark (n = 10). (D) Kinetics of 8-oxoG at sites of tetR-KR or KR-TRF1 transfected U2OS (TRE) cells with or without light activation of KR, recovered in the dark for the indicated time. (E) Quantification of the intensity of 8-oxoG at sites of tetR-KR or KR-TRF1 (n = 10) at the indicated time point after light activation of KR for 20 min.
