Figure 1.
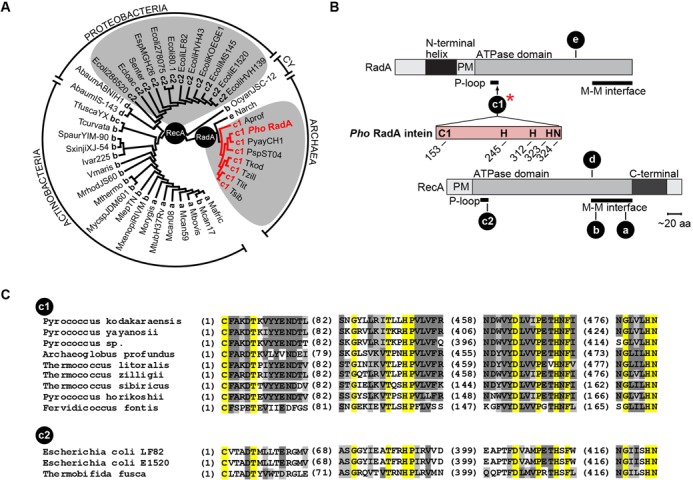
Distribution and diversity of RadA/RecA inteins among archaea and bacteria. (A) Neighbor-Joining tree. The tree was constructed based on the multiple alignment of 42 RadA/RecA extein amino acid sequences from archaea and bacteria (proteobacteria, actinobacteria and cyanobacteria—CY). The insertion points (a–e) are indicated together with abbreviated species names, listed in full in Table 1. Letters a–e reflect the chronology of intein discovery (54,55). The species containing an intein in insertion point c (either c1 or c2) are on a gray background. The precursor of the protein of interest, Pho RadA (bold, red), carries an intein in insertion point c, subpopulation c1. (B) Intein insertion points. Intein locations are shown along RadA (c1 and e) and RecA (a–d) relative to structural and functional domains. The intein of interest, Pho RadA, is in insertion point c1 (red asterisk), which is located at the end of the RadA P-loop in the ATPase domain. Conserved, catalytically important amino acid residues and their positions are shown in single-letter code in the intein, as further identified in Figure 4A. Abbreviations: PM, polymerization motif; M–M interface, monomer–monomer interface. (C) Multiple sequence alignment of the inteins (splicing domains) from insertion point c. Comparative analysis of the splicing domains from inteins occupying insertion point c in RadA and RecA proteins revealed substantial differences in amino acid sequences between archaeal (insertion c1) and bacterial (insertion c2) inteins indicating independent acquisition of inteins by RadA and RecA in insertion point c.
