Figure 1.
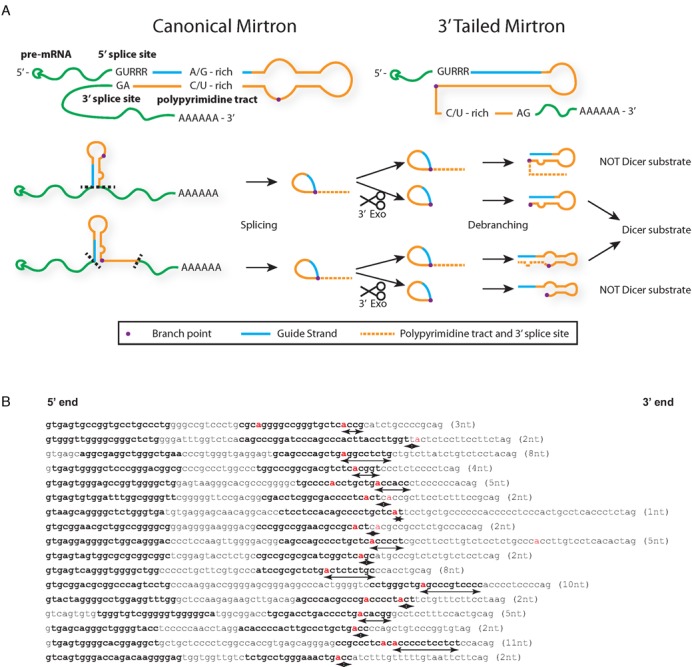
Proposed pathways for biogenesis of mirtrons and tailed mirtrons. (A) Canonical mirtrons have strong sequence constraints due to the need to incorporate splicing motifs within the hairpin sequence for spliceosome recognition, including the 5′ splice site (GURRR), the purine-rich 5′ arm (A/G-rich) to complement the polypyrimidine tract (C/U-rich), while tailed mirtrons are only constrained by the 5′ splice site sequence. The sequence of events following splicing is still not clear currently; thus, the interplay between 3′ exonuclease degradation by the exosome (dotted line represents sequence that is degraded) and debranching can result in different hairpins that may or may not be Dicer substrates. (B) Identified human 3′-tailed mirtrons from reference 13 (http://ericlailab.com/mammalian_mirtrons/hg19/). Only genuine 3′-tailed mirtrons with a 5′ arm less than 10 nt from the 5′ splice site (to exclude intronic miRNAs) and a branch point (red) that can be predicted by a branch point prediction algorithm (14) were included. Bold letters represent annotated guide and passenger strands from reference 13. Distance from the branch point to the 3′ end of the hairpin indicated in parentheses and in arrows under the sequence.
