Figure 2.
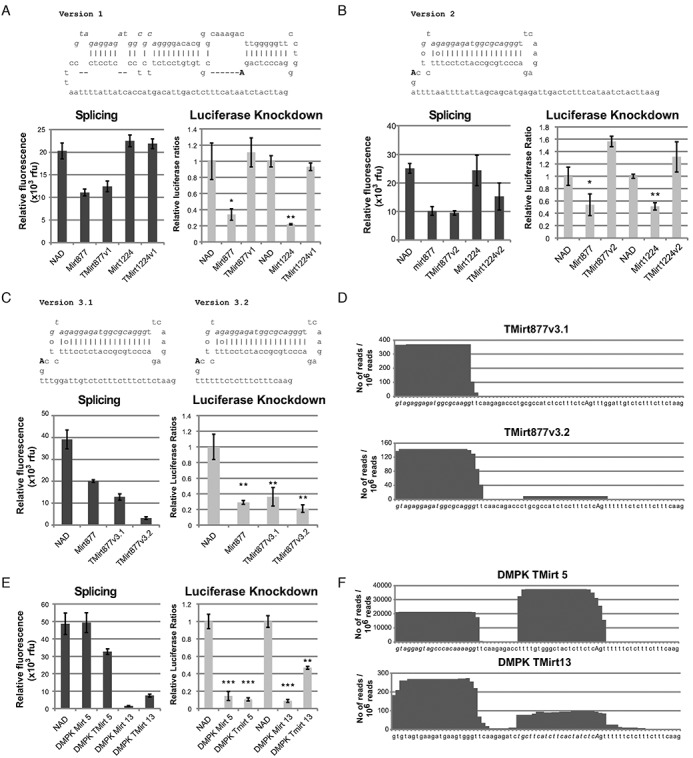
Development of effective 3′-tailed mirtrons. HEK cells were transfected with 100 ng of psiCheck2.2 targets, with target sequences in the 3′ UTR of Renilla luciferase, and 500 ng of mirtron plasmid, and lysed two days later for fluorescence measurement and luciferase activity. (A–C) eGFP fluorescence quantification, Renilla luciferase knockdown and different TMirt designs with hsa-mir-877 and mmu-mir-1224 guide strands in a pEGFP-Mirt vector. Guide strands and putative branch points are marked by italic and uppercase text respectively. (D) High-throughput sequencing was performed with 18–25nt RNA from HEK293 cells transfected with TMirt877v3.1 and v3.2. Sequences that were aligned to mirtron sequences were collected, 3′ tail removed and aligned to chart the frequency of individual nucleotides appearing in the small RNA species harvested from the cells. Nucleotides in italics denote guide strand, uppercase denotes branch point. (E) TMirts based on canonical mirtrons designed against DMPK were compared to the parent mirtrons based on splicing efficiency (eGFP fluorescence) and Renilla luciferase knockdown. (F) High-throughput sequencing of small RNAs from HEK293 cells transfected with 500 ng of the relevant plasmids. *P < 0.05, **P < 0.01, ***P < 0.001 versus NAD control, n = 3 biological replicates for each experiment. Error bars reflect standard deviation.
