Figure 4.
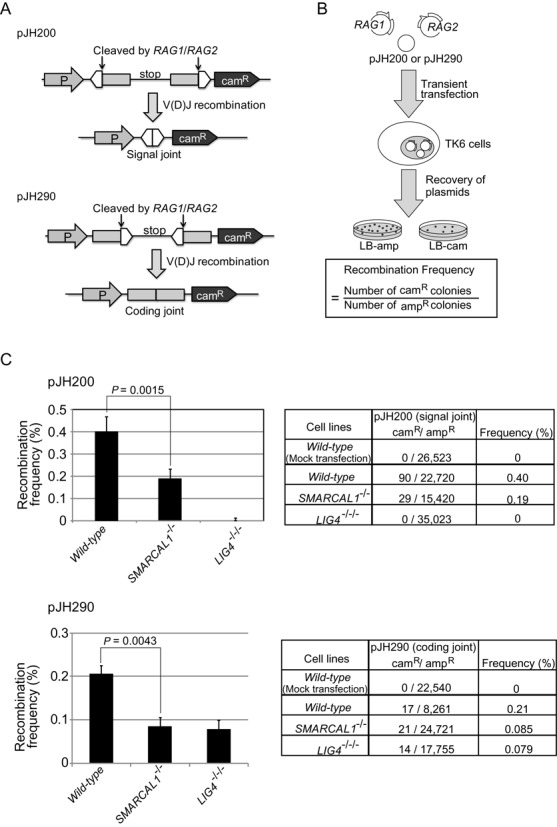
The loss of Smarcal1 reduces the efficiency of V(D)J recombination without compromising its fidelity. (A) The structure of two episomal V(D)J-recombination substrates, pJH200 and pJH290, and their recombination products. Open triangles and closed boxes represent recombination signals (RSs) and V(D)J-coding sequences, respectively. CamR = chloramphenicol-resistance gene; P = promoter. (B) Schematic representation of the experimental method for the V(D)J-recombination assay. Frequency of recombination was calculated by dividing the number of rearranged products (the number of camR colonies) by the number of recovered plasmids (the number of ampicillin-resistant [ampR] colonies). (C) Recombination frequency of TK6 cells carrying the indicated genotypes. Data shown are the means of more than three experiments. Error bars indicate SD of more than three independent experiments. P-value was calculated by Student's t-test. The total number of ampicillin- and chloramphenicol-resistant colonies is shown in the right panel. Supplemental Table S2 shows the nucleotide sequences of coding joints associated with deletion events.
