Figure 4.
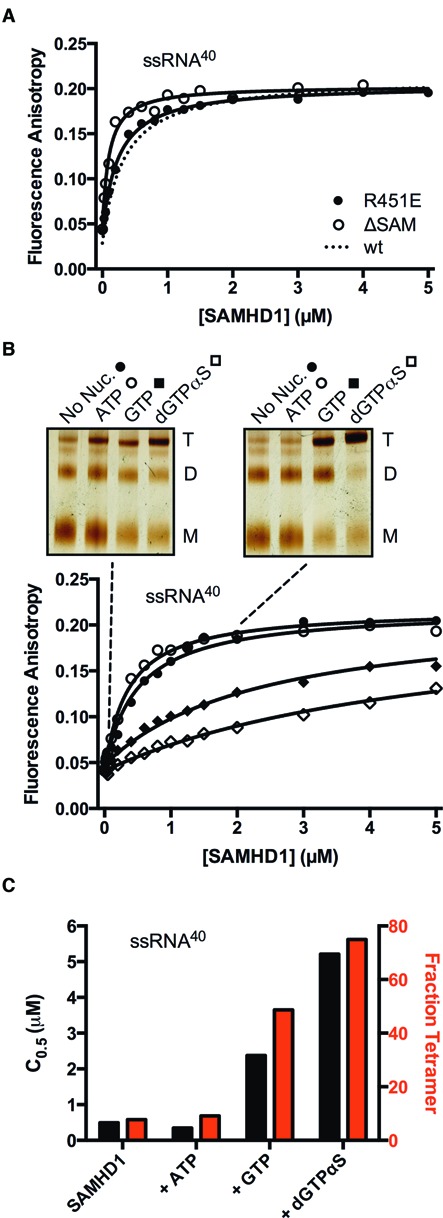
Binding of SAMHD1 to ssRNA40 does not require the SAM domain and is inhibited by guanine nucleotide-induced protein tetramerization. (A) Fluorescence anisotropy changes accompanying binding of the ΔSAM deletion mutant and the monomeric R451E mutant to ssRNA40 (10 nM). The binding curve for wt-SAMHD1 was very similar under the same conditions and is shown for comparison (dashed line). (B) Binding of wt-SAMHD1 to ssRNA40 was measured in the absence of nucleotide, and in the presence of ATP (non-activator), GTP, or dGTPαS (all at 1 mM concentration). For each nucleotide condition, the oligomeric state of SAMHD1 at two concentrations is indicated with the dashed lines (0.1 and 2 μM), which was determined by glutaraldehyde crosslinking and SDS-PAGE (insets). (C) The C0.5 values (black bars) and fraction of total SAMHD1 in the tetrameric form (red bars) for each nucleotide condition are correlated.
