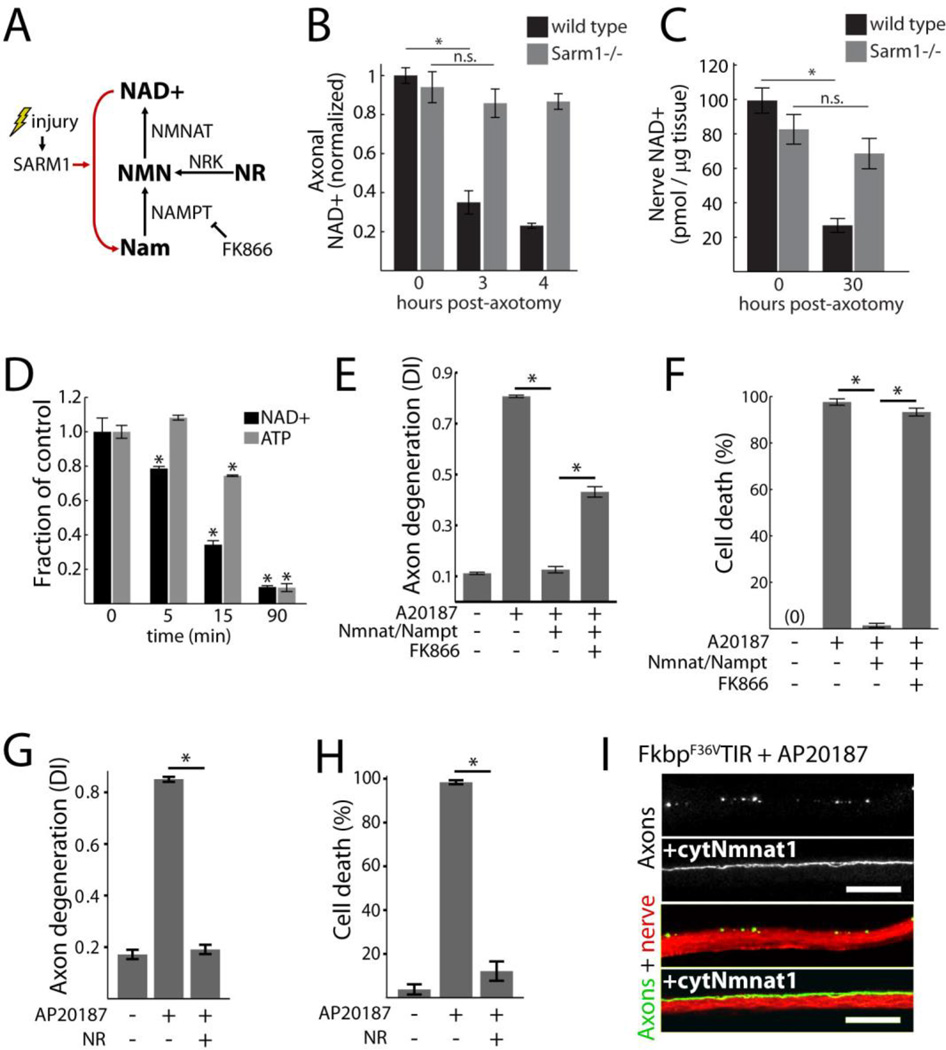
Fig. 3.
Loss of NAD+ underlies SARM1-induced destruction. A) Diagram of NAD+ synthesis and inhibition by FK866; Nam=nicotinamide, NR = nicotinamide riboside, Nrk=nicotinamide riboside kinase, NMN=nicotinamide mononucleotide. B) Axonal NAD+ concentration in cultured wild-type and Sarm1−/− DRG neurons following axotomy; normalized to wild-type control. C) NAD+ concentration in distal sciatic nerve segments from wild-type or Sarm1−/− animals following transection; wild-type n=5; Sarm1−/− n=9. D) Neuronal NAD+ and ATP concentrations following sTIR dimerization by AP20187; comparisons made to zero minutes control. E–F) Axon degeneration (E) and neuronal cell death (F) induced by sTIR homodimerization (AP20187) and inhibition by NAD+ synthetic enzymes with or without the Nampt inhibitor FK866 (10 nM); measured 24 hours after sTIR dimerization and FK866 application. G–H) Effect of NR (1 mM) on axon degeneration (G) and neuronal cell death (H) induced by sTIR homodimerization (AP20187) for 24 hours with or without NR. I) Micrographs showing sTIR-induced motor axon fragmentation in third instar Drosophila larvae blocked by cytosolic Nmnat1 (cytNmnat1) expression. M12-Gal4 drives expression from UAS-mCD8-GFP (green) and UAS-FkF36VTIR with or without UAS-cytNmnat1 in single motor axons in each nerve (red=HRP). Degeneration score = 76+/−4% (control) vs 11+/−2% (cytNmnat1); p<0.001 (t-test); scale bar=20 micrometers. Error bars = SEM; * p < 0.01; one-way ANOVA with Tukey’s post-hoc test.