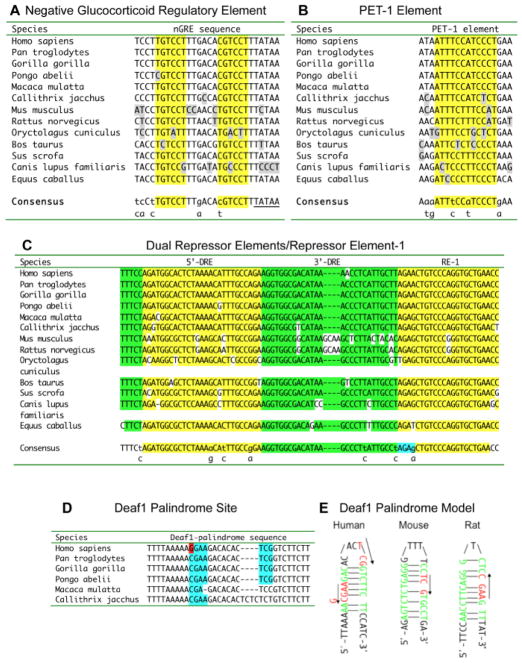
Fig. 5. Conserved HTR1A DNA elements.
The alignment of DNA elements from mammalian 5-HT1A receptor gene sequences is shown with elements of interest highlighted; below is the consensus sequence shared in three or more species; mismatches from the consensus are shown in grey.
A. Negative Glucocorticoid Regulatory Element (nGRE). The GRE half-site sequence is highlighted and a 3′-flanking TATA-box sequence of unknown function is also shown. B. PET-1 element. C. Dual repressor elements (DRE)/repressor element-1 (RE-1). The 5′DRE and RE-1 are highlighted in yellow; 3′-DRE and conserved upstream sequence in green; overlap in blue; and mismatches in white. D. Deaf1 palindrome site. Shown are conserved sequences in primate species; the minimal Deaf1 recognition sequence is in blue; the site of the rs6295 polymorphism (G-allele shown) is in red. E. Deaf1 palindrome model. Deaf1 binding sites in human, mouse and rat 5-HT1A promoters are shown as “hybridized” to highlight imperfect palindrome sequences shown in green; the consensus Deaf1 minimal TCG (forward) or CGA (reverse) is shown in red. The location of the G(-1019) rs6295 site in the human 5-HT1A palindrome is indicated.