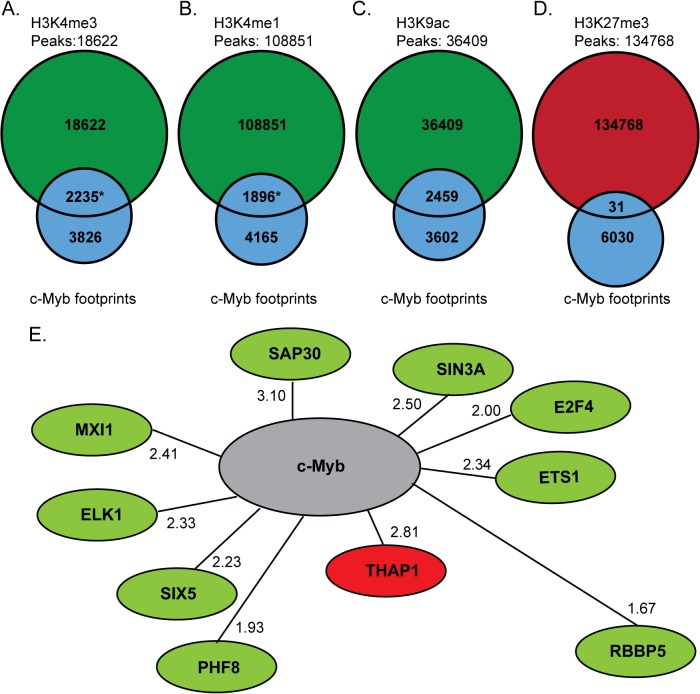
Fig 4. c-Myb footprints co-localises with active histone marks and co-regulatory TFs at c-Myb-regulated genes.
(A-D) The number of c-Myb footprints (grey), which co-localize with ChIP-seq peaks for the active histone marks H3K4me3, H3K4me1, H3K9ac, (green) and the repressive mark H3K27me3 (red) in K562 cells. *Significantly different than expected from random sampling K562 DNase I footprints (p', > 4x10-4). (E) Suggested co-regulatory TFs for c-Myb in K562 cells. Green and red denotes factors in the positive and negative set respectively. Next to each factor, the normalised ratio for the co-localisation with c-Myb footprints at positively or negatively regulated genes is displayed. The distance between each factor and c-Myb is calculated as a measure of the normalized ratio, as described in Methods.