Abstract
Candidatus Neoehrlichia mikurensis (CNM; family Anaplasmataceae) was recently recognized as a potential tick-borne human pathogen. The presence of CNM in mammals, in host-seeking Ixodes ticks and in ticks attached to mammals and birds has been reported recently. We investigated the presence of CNM in ornithophagous ticks from migrating birds. A total of 1,150 ticks (582 nymphs, 548 larvae, 18 undetermined ticks and two adult females) collected from 5,365 birds captured in south-eastern Sweden was screened for CNM by molecular methods. The birds represented 65 different species, of which 35 species were infested with one or more ticks. Based on a combination of morphological and molecular species identification, the majority of the ticks were identified as Ixodes ricinus. Samples were initially screened by real-time PCR targeting the CNM 16S rRNA gene, and confirmed by a second real-time PCR targeting the groEL gene. For positive samples, a 1260 base pair fragment of the 16S rRNA gene was sequenced. Based upon bacterial gene sequence identification, 2.1% (24/1150) of the analysed samples were CNM-positive. Twenty-two out of 24 CNM-positive ticks were molecularly identified as I. ricinus nymphs, and the remaining two were identified as I. ricinus based on morphology. The overall CNM prevalence in I. ricinus nymphs was 4.2%. None of the 548 tested larvae was positive. CNM-positive ticks were collected from 10 different bird species. The highest CNM-prevalences were recorded in nymphs collected from common redpoll (Carduelis flammea, 3/7), thrush nightingale (Luscinia luscinia, 2/29) and dunnock (Prunella modularis, 1/17). The 16S rRNA sequences obtained in this study were all identical to each other and to three previously reported European strains, two of which were obtained from humans. It is concluded that ornithophagous ticks may be infected with CNM and that birds most likely can disperse CNM-infected ticks over large geographical areas.
Introduction
Candidatus Neoehrlichia mikurensis (CNM) is a recently recognized tick-transmitted potential human pathogenic rickettsia of the family Anaplasmataceae. This obligately intracellular gram-negative bacterium is transmitted to humans by ticks. It was first described after being found in Ixodes ovatus and isolated from brown rats (Rattus norvegicus) in Japan in 2004 [1]. However, it was later realized that this bacterium had previously been detected in I. ricinus in the Netherlands but was at that time referred to as an Ehrlichia-like species [2]. In 2010, the first human CNM-infected patient was described in Sweden [3]. So far, 14 human cases have been reported in Europe, predominantly in immunocompromised patients [4–6]. Another seven human cases have been reported in China, all in previously healthy individuals [7]. The symptoms include persistent fever, chills, arthralgia, myalgia, headache and thromboembolic events. One patient died from the CNM infection [8]. Diagnosis is currently based on molecular detection of the CNM 16S rRNA gene in blood [9], since no isolation technique has been established.
In the last years, studies conducted in different European countries have shown that the prevalence of CNM in field-collected host-seeking I. ricinus ticks may range from 0% to 12% [10]. The CNM prevalence in The Netherlands was reported as 6, 8, 4 and 13% in adult I. ricinus removed from red deer (Cervus elaphus), wild boar (Sus scrofa), sheep (Ovis aries) and mouflon (Ovis orientalis musimon), respectively [8]. CNM was found in 5 of 276 (1.8%) spleens from bank voles (Myodes glareolus) in France [11]. In southern Sweden, the prevalence of CNM was 9.0% in M. glareolus and 5.7% in Apodemus spp. [12]. These studies show that CNM is present in both I. ricinus and in mammals in Europe. Given the fact that I. ricinus is commonly retrieved from wild birds, it is important to study the potential role of birds in the epidemiology of CNM. Hypothetically, birds may be competent transmission hosts for CNM or incompetent transmission hosts but yet hosts of ticks, which could be CNM-infected. In both ways it could be conceivable that birds can spread the bacterium to new regions along their migratory flyways [13, 14]. The first record of CNM in a tick infesting a bird is from 2006, when Spitalska et al. detected an Ehrlichia-like species, referred to as the “Schotti variant”, in an I. ricinus nymph collected from a song thrush (Turdus philomelos) in Slovakia [15]. In 2012, Movila et al. screened ticks from migratory birds in the Baltic Region of Russia; CNM was found in 1 of 135 (0.7%) I. ricinus and in 1 of 4 I. frontalis [16]. In a recent study from Switzerland, CNM was found in 7 of 215 ticks (3.3%) collected from breeding and migratory birds. All seven CNM-positive ticks were I. ricinus nymphs. Six of them were collected from four migrant chaffinches (Fringilla coelebs) and one nymph was collected from a wren (Troglodytes troglodytes) [14].
These studies are limited, but suggest that CNM can be found in ticks feeding on birds. In the present study, a large representative tick material retrieved from birds caught during stopover on spring and autumn migration in South Sweden was screened for CNM. Combining molecular screening for CNM and molecular species identification of ticks, we aimed to provide a better baseline for the presence of CNM in birds. If birds act as transmission hosts of CNM to blood-feeding ticks, we hypothesize that CNM should be detected in all active stages of strictly ornitophagous (bird-feeding) ticks and in tick larvae of species with a broader host range retrieved from birds.
Materials and Methods
Bird trapping
All birds were trapped during the normal trapping activities of staff members at the Ottenby Bird Observatory (56° 12′ N, 16° 24′ E), Sweden, under a general ringing license from the Swedish Ringing Office (Ringmärkningscentralen, Naturhistoriska riksmuseet) in accordance with Swedish regulations. Sampling of birds was approved by the Swedish Board of Agriculture, delegated through Linköpings Djurförsöksetiska nämnd in Linköping (decision 43–09).
Tick collection
During 2009, 5,365 migratory birds caught at Ottenby Bird Observatory were examined for ticks around their bills and eyes and in the ears. The site of attachment on the bird was noted for each tick before it was removed, photographed for identification of species and stage, and stored at -70°C until further investigation. In total, 1,335 ticks from 748 birds were collected. Of these, 21 ticks could not be removed from the birds and 162 ticks were lost due to technical problems during nucleic acid extraction, resulting in 1,150 ticks available for analysis.
Tick identification
Each tick was photographed dorsally and ventrally using a digital USB-microscope, Dino-Lite Long AM4013TL (AnMo Electronics Corp., Taiwan). Based on photographs and the descriptions in [17–20], the ticks were morphologically identified, i.e. the developmental stage (larva, nymph or adult), sex of adults and genus of larvae, nymphs and adults were recorded. The degree of blood ingested by each tick was estimated as unfed (U), little fed (LF; a relatively small amount of host blood in the tick's gut), half fed (HF) or fully fed (FF). The digital photographs did not always capture the distinguishing criteria, or the tick had been damaged during removal from the birds. Determination of the species was therefore often uncertain or even impossible. To remedy this, all nymphs and adults were further screened by molecular methods. The cDNA from the CNM screening was used as template in the PCR reactions. The first method targeted the mitochondrial gene cytochrome C oxidase subunit I (COI), which was amplified by the primers Cox1F and Cox1R (Table 1) [21]. The PCR reactions were carried out in 25 μl reaction volumes, containing 2 μl of template cDNA, 10 μM of forward and reverse primers, 5 μl Q-solution, 2.5 μl 1x PCR buffer (Applied Biosystems, The Netherlands), 0.2mM deoxynucleoside triphosphates, 1.5mM MgCl2 and 1.25U of Amplitaq polymerase (Applied Biosystems, The Netherlands). Thermal conditions included incubation at 94°C for 3 min followed by 45 cycles at 95°C for 30 s, 45°C for 30 s and 72°C for 3 min, and a final 7 min elongation at 72°C. Samples that could not be identified with the COI method were further analysed with a method that targeted the mitochondrial 16S rDNA gene. The gene was amplified using the forward primer m16sFW/tck and the reverse primer m16sRev/tck (Table 1) [20]. The PCR reactions were carried out in 25 μl reaction volumes, containing 5ul of template cDNA, 10 μM of forward and reverse primers, 5 μl Q-solution, 2.5 μl 1x PCR buffer (Applied Biosystems, The Netherlands), 0.2mM deoxynucleoside triphosphates, 1.5mM MgCl2 and 1.25U of Amplitaq polymerase (Applied Biosystems, The Netherlands). PCR was performed with an initial denaturation step at 94°C for 4 min followed by 35 cycles of 30 s at 92°C, 30 s at 50°C and 45 s at 72°C and a final extension step for 8 min at 72°C. Thermal conditions included incubation at 94°C for 4 min, followed by 35 cycles at 92°C for 30 s, 50°C for 30 s and 72°C for 45 s, and a final 8 min elongation at 72°C. Positive samples were sequenced with the forward primer at a commercial centre (Macrogen Europe, The Netherlands).
Table 1. Primers used for screening and sequencing.
| Primer name | Target gene | Primer sequence | Reference |
|---|---|---|---|
| Neo_16S_F | 16S rRNA | 5'-GTAAAGGGCATGTAGGCGGTTTAA-3' | Primers were established as part of this study |
| Neo_16S_R | 16S rRNA | 5'-TCCACTATCCTCTCTCGATCTCTAGTTTAA-3' | Primers were established as part of this study |
| Neo_16S_95_F | 16S rRNA | 5'-TTAGTGGCAGACGGGTGAGTAATG-3' | Primers were established as part of this study |
| Neo_16S_127_F | 16S rRNA | 5'-TCTGCCTAGTAGTATGGAATAGCTG-3' | Primers were established as part of this study |
| Neo_16S_1363_R | 16S rRNA | 5'-AAACCAATTTCCAGGGCATGACGG-3' | Primers were established as part of this study |
| Neo_16S_1393_R | 16S rRNA | 5'-TCCTTACGGTTAGCTCACCAGCTT-3' | Primers were established as part of this study |
| NeogroELQf | GroEL | 5'-ACAGCCAATACTACCTATCCTTGA-3' | Primers, although with a slightly modified version of the reverse primer, were initially reported by Andersson and colleagues [22] |
| NeogroELQr | GroEL | 5'-ACATGTAATCCACCACGCAACT-3' | Primers, although with a slightly modified version of the reverse primer, were initially reported by Andersson and colleagues [22] |
| Cox1F | COI | 5´-GGAACAATATATTTAATTTTTGG-3´ | [21] |
| Cox1R | COI | 5´-ATCTATCCCTACTGTAAATATATG-3´ | [21] |
| m16sFW/tck | 16S rRNA | 5´-CCGGTCTGAACTCAGATCAAGT-3´) | [20] |
| m16sRev/tck | 16S rRNA | 5´-GCTCAATGATTTTTTAAATTGCTGT-3´) | [20] |
Molecular CNM detection, sequencing and phylogenetic analysis
Extracting total nucleic acid (DNA and RNA) from a tick allows for the monitoring of known and emerging vector-borne pathogens. Therefore, after total nucleic acid extraction and cDNA synthesis as previously described [23], cDNA was screened using a real-time PCR protocol including specific primers targeting the CNM 16S rRNA-gene (Table 1) to generate 107 bp DNA fragments using the 2X iQ SYBR Green supermix (Bio-Rad Laboratories, USA) on either a StepOne Plus (Applied Biosystems, The Netherlands) or a Light Cycler 480 (Roche, Switzerland) instrument. Thermal cycling conditions included an initial denaturation step at 95°C for 3 min, followed by 45 cycles of 95°C for 15 s, 60°C for 30 s and 72°C for 30 s. In addition, a melting curve analysis of amplified products was performed between 55°C and 95°C. Cycle threshold (Ct) values below 40 and melting temperatures between 78.5 and 79.2°C were considered as CNM positive.
In order to further validate signals in positive samples, cDNA determined positive by detection of the 16S rRNA gene was examined using a real-time PCR protocol targeting the CNM groEL gene using previously described methods [22].
For phylogenetic analysis, a 1262 bp of the CNM 16S rRNA gene was sequenced, employing a nested PCR method including specific primers (Table 1). Thermal conditions included incubation at 94°C for 5 min, followed by 45 cycles at 95°C for 30 s, 58°C (PCR1) or 55°C (PCR2) for 30 s and 72°C for 3 min, and a final 7 min elongation at 72°C. Amplifications were performed using the Expand High FidelityPLUS PCR System (Roche, Switzerland) according to the manufacturer’s protocol, by using 2 μl of cDNA sample for the first PCR, and by diluting the resulting PCR product 100 times. One μl of this dilution was used as template in the second PCR. Amplified products were purified and sequenced by the Sanger method (Macrogen Europe, The Netherlands).
Results and Discussion
A total of 5,365 birds were captured during the ringing season of 2009. Some birds were caught two or more times and each capture was counted separately. The captured birds represented 65 different species, of which 35 species were infested with one or more ticks. The most commonly infested bird species were tree pipit (Anthus trivialis, 87.7%), thrush nightingale (Luscinia luscinia, 78.4%) and common blackbird (Turdus merula, 64.8%) (Table 2). Of the 1,150 ticks that were screened for CNM, two were adult females, 582 were nymphs, 548 were larvae and 18 were undetermined (Table 3). Using morphological identification, 1,110 ticks were determined as Ixodes spp., 12 as Haemaphysalis spp. and 4 as Hyalomma spp. Genus identification was not possible in 24 ticks (Table 3). Since CNM was recorded only from nymphs, all nymphs and the two adult female ticks were further analysed by molecular methods. A total of 569 ticks (2 adults and 567 nymphs) were identified using COI. Of the analysed nymphs, 526 were identified as I. ricinus, five as I. arboricola, two as Hyalomma marginatum and four as Ixodes spp. Another 30 nymphs either did not amplify or did not give readable sequences despite several sequencing attempts. Thirty-nine ticks that could not be identified by COI were further screened using the 16S rRNA PCR. This method identified 19 I. ricinus and 8 I. frontalis ticks. Six nymphs were identified as Ixodes spp. Four nymphs and one adult female could not be identified either by COI or 16S rRNA PCR. The different results in the morphological and the molecular tick identification methods might be due to several reasons. As mentioned before, morphological identification of ticks from photographs is associated with uncertainties. Lv et al. developed a DNA barcoding system for Ixodida in 2013 [24] and they conclude that there still are some serious deficiencies in the information of 16S and COI of some species of ticks. Furthermore, the performance of DNA barcoding can be influenced if species previously submitted to GenBank are incorrectly identified [25].
Table 2. Prevalence of tick infestation and Candidatus Neoehrlichia mikurensis in migratory birds captured at Ottenby, Sweden in 2009.
| Bird species | No of birds examined for ticks* | Number of birds with tick | % of birds infested with ticks | Number of ticks screened for CNM | Number of CNM-positive ticks | % CNM-positive ticks |
|---|---|---|---|---|---|---|
| Accipiter nisus | 5 | 1 | 20.0 | 1 | 0 | 0 |
| Acrocephalus arundinaceus | 1 | 0 | 0 | 0 | 0 | 0 |
| Acrocephalus palustris | 22 | 4 | 18.2 | 4 | 0 | 0 |
| Acrocephalus schoenobaenus | 23 | 0 | 0 | 0 | 0 | 0 |
| Acrocephalus scirpaceus | 11 | 1 | 9.1 | 1 | 0 | 0 |
| Anthus pratensis | 5 | 0 | 0 | 0 | 0 | 0 |
| Anthus trivialis | 106 | 93 | 87.7 | 85 | 1 | 1.2 |
| Carduelis cannabina | 15 | 1 | 6.7 | 1 | 0 | 0 |
| Carduelis carduelis | 15 | 0 | 0 | 0 | 0 | 0 |
| Carduelis chloris | 16 | 0 | 0 | 0 | 0 | 0 |
| Carduelis flammea | 41 | 7 | 17.1 | 7 | 3 | 42.9 |
| Carduelis spinus | 9 | 0 | 0 | 0 | 0 | 0 |
| Carpodacus erythrinus | 2 | 0 | 0 | 0 | 0 | 0 |
| Certhia familiaris | 28 | 4 | 14.3 | 4 | 0 | 0 |
| Coccothraustes coccothraustes | 1 | 0 | 0 | 0 | 0 | 0 |
| Corvus monedula | 1 | 0 | 0 | 0 | 0 | 0 |
| Delichon urbica | 43 | 0 | 0 | 0 | 0 | 0 |
| Dendrocopus major | 1 | 0 | 0 | 0 | 0 | 0 |
| Emberiza citrinella | 23 | 0 | 0 | 0 | 0 | 0 |
| Emberiza schoeniclus | 29 | 3 | 10.3 | 1 | 0 | 0 |
| Erithacus rubecula | 1796 | 619 | 34.4 | 513 | 6 | 1.2 |
| Ficedula albicollis | 2 | 0 | 0 | 0 | 0 | 0 |
| Ficedula hypoleuca | 31 | 0 | 0 | 0 | 0 | 0 |
| Ficedula parva | 8 | 0 | 0 | 0 | 0 | 0 |
| Fringilla coelebs | 39 | 8 | 20.5 | 8 | 0 | 0 |
| Fringilla montifringilla | 8 | 1 | 12.5 | 1 | 0 | 0 |
| Hippolais icterina | 50 | 2 | 4.0 | 2 | 0 | 0 |
| Hirundo rustica | 25 | 0 | 0 | 0 | 0 | 0 |
| Jynx torquilla | 1 | 0 | 0 | 0 | 0 | 0 |
| Lanius collurio | 43 | 3 | 7.0 | 3 | 0 | 0 |
| Locustella naevia | 1 | 0 | 0 | 0 | 0 | 0 |
| Luscinia luscinia | 37 | 29 | 78.4 | 29 | 2 | 6.9 |
| Luscinia svecica | 28 | 2 | 7.1 | 1 | 0 | 0 |
| Motacilla alba | 25 | 3 | 12.0 | 3 | 0 | 0 |
| Motacilla flava | 2 | 0 | 0 | 0 | 0 | 0 |
| Muscicapa striata | 55 | 0 | 0 | 0 | 0 | 0 |
| Oenanthe oenanthe | 6 | 1 | 16.7 | 1 | 0 | 0 |
| Parus caeruleus | 61 | 7 | 11.5 | 7 | 0 | 0 |
| Parus major | 49 | 8 | 16.3 | 2 | 0 | 0 |
| Passer domesticus | 16 | 1 | 6.3 | 1 | 0 | 0 |
| Passer montanus | 37 | 0 | 0 | 0 | 0 | 0 |
| Phoenicurus ochruros | 10 | 2 | 20.0 | 3 | 0 | 0 |
| Phoenicurus phoenicurus | 157 | 64 | 40.8 | 61 | 1 | 1.6 |
| Phylloscopus borealis | 1 | 0 | 0 | 0 | 0 | 0 |
| Phylloscopus collybita | 79 | 3 | 3.8 | 2 | 0 | 0 |
| Phylloscopus sibilatrix | 19 | 0 | 0 | 0 | 0 | 0 |
| Phylloscopus trochilus | 829 | 57 | 6.9 | 54 | 0 | 0 |
| Picus viridis | 1 | 0 | 0 | 0 | 0 | 0 |
| Prunella modularis | 48 | 20 | 41.7 | 17 | 1 | 5.9 |
| Pyrrhula pyrrhula | 2 | 0 | 0 | 0 | 0 | 0 |
| Regulus regulus | 319 | 9 | 2.8 | 8 | 0 | 0 |
| Saxicola rubetra | 1 | 0 | 0 | 0 | 0 | 0 |
| Serinus serinus | 2 | 0 | 0 | 0 | 0 | 0 |
| Sturnus vulgaris | 14 | 3 | 21.4 | 3 | 0 | 0 |
| Sylvia atricapilla | 58 | 4 | 6.9 | 4 | 0 | 0 |
| Sylvia borin | 42 | 0 | 0 | 0 | 0 | 0 |
| Sylvia communis | 145 | 49 | 33.8 | 44 | 1 | 2.3 |
| Sylvia curruca | 271 | 11 | 4.1 | 10 | 0 | 0 |
| Sylvia nisoria | 1 | 0 | 0 | 0 | 0 | 0 |
| Troglodytes troglodytes | 301 | 120 | 39.9 | 95 | 2 | 2.1 |
| Turdus iliacus | 42 | 23 | 54.8 | 21 | 1 | 4.8 |
| Turdus merula | 230 | 149 | 64.8 | 133 | 6 | 4.5 |
| Turdus philomelos | 73 | 22 | 30.1 | 20 | 0 | 0 |
| Turdus pilaris | 1 | 0 | 0 | 0 | 0 | 0 |
| Turdus viscivorus | 1 | 0 | 0 | 0 | 0 | 0 |
| Unknown (probably Lanius collurio) | 1 | 1 | 100.0 | 0 | 0 | 0 |
| Total | 5365 | 1335 | 24.9 | 1150 | 24 | 2.1 |
Table 3. CNM prevalence in different tick genera and tick stages.
Genus identification is based on morphological identification. The numbers represent total number of ticks/ticks screened for CNM/CNM-positive ticks.
| Adult female | Nymphs | Larvae | Un-determined | Total all stages | Total CNM-positive | CNM prevalence | |
|---|---|---|---|---|---|---|---|
| Haemaphysalis spp. | 0 | 1/1/0 | 11/11/0 | 0 | 12/12/0 | 0 | 0% |
| Hyalomma spp. | 1/1/0 | 2/2/0 | 2/1/0 | 0 | 5/4/0 | 0 | 0% |
| Ixodes spp. | 1/1/0 | 613/579/24 | 589/528/0 | 3/2/0 | 1206/1110/24 | 24 nymphs | 2.2% |
| N/A | 0 | 3/0/0 | 11/8/0 | 98/16/0 | 112/24/0 | 0 | 0% |
| Total number of ticks | 2/2/0 | 619/582/24 | 613/548/0 | 101/18/0 | 1335/1150/24 | 24 nymphs | 2.1% |
| CNM prevalence | 0% | 4.1% | 0% | 0% |
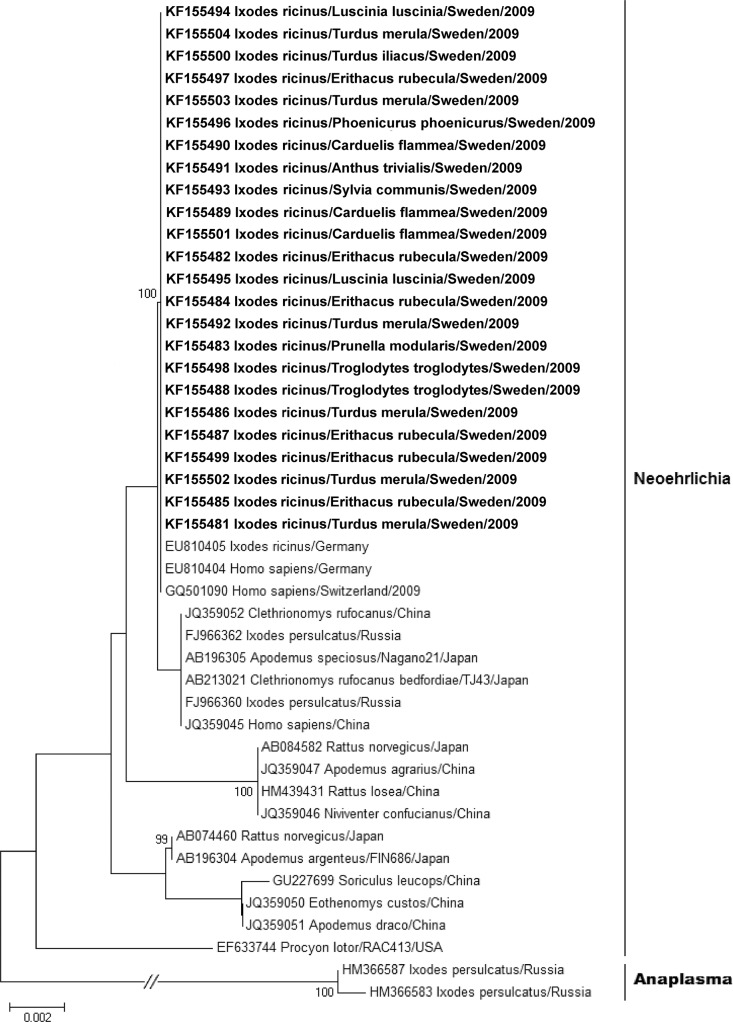
Forty-nine ticks (i.e. 4.3%) were positive for CNM when screened with the 16S-based PCR method, and 31 of these remained positive after the groEL-PCR screening. Sequencing revealed that the established 16S rRNA-PCR lacked somewhat in selectivity since it also detected bacteria other than CNM, such as Rickettsia spp., Candidatus Midichloria spp. and others. However, CNM sequences were obtained from 24 of the 31 samples that were positive in screenings with both the 16S and the groEL method. Alignment and phylogenetic analysis [26] showed that the CNM sequences were identical to each other as well as to three other European strains, including those detected in humans, and appeared in a single clade (Fig 1). Thus, based on bacterial gene sequence identification, 2.1% (24/1150) of analysed ticks were concluded to be CNM positive. This might be an underestimation of the true prevalence, since the CNM RNA level in the samples could be too low to be detected.
Fig 1. Phylogenetic relatedness of Candidatus Neoehrlichia mikurensis sequences.
The tree is based on the partial nucleotide sequence of the 16S rRNA gene of CNM and Anaplasma phagocytophilum. Bacterial sequences generated in this study (boldface) and previously reported sequences are denoted with GenBank accession number followed by information regarding sequence source. The tree was inferred using the neighbour joining method implemented in the SeaView software version 4 [27], utilizing 1000 bootstrap replications to determine support for inferred nodes. The tree was visualized using the MEGA software version 5.2 [28], and statistical supports of > 95% for inferred nodes are displayed. In the mid-point rooted tree, the branch between the CNM and the Anaplasma clade has been truncated in order to limit the tree size. The scale bar represents the number of substitutions per site. The CNM sequences were all 100% identical to each other as well as to three other European strains, including those detected in humans, and therefore appear in a single clade.
According to molecular species identification, I. ricinus was the dominant tick species collected from the birds in this study. The CNM-infected ticks were all nymphs and 22 of them were identified as I. ricinus (Table 4). The remaining two CNM-infected ticks were morphologically identified as I. ricinus, but were not subjected to molecular testing. The prevalence of CNM in molecularly identified I. ricinus nymphs was 4.2% (22/526) (Table 3). None of the 548 larvae was positive for CNM.
Table 4. Candidatus Neoehrlichia mikurensis-positive tick nymphs from birds in Sweden 2009.
| Bird no | Bird species | Migration | Collection month | Tick species § | Feeding status | Tick location | |
|---|---|---|---|---|---|---|---|
| Spring | 1 | Turdus merula | SD* | March | I. ricinus | HF | Bill |
| 2 | Turdus merula | SD | April | I. ricinus | LF | Bill | |
| 3 | Anthus trivialis | SD | April | I. ricinus | HF | Bill | |
| 4 | Erithacus rubecula | SD | April | I. ricinus | FF | Bill | |
| 5 | Prunella modularis | SD | April | I. ricinus | U | Bill | |
| 6 | Erithacus rubecula | SD | April | I. ricinus | FF | Bill | |
| 7 | Erithacus rubecula | SD | April | I. ricinus | FF | Bill | |
| 8 | Erithacus rubecula | SD | April | I. ricinus | LF | Bill | |
| 9† | Carduelis flammea | SD | April | I. ricinus | LF | Eye | |
| 9† | Carduelis flammea | SD | April | I. ricinus | HF | Eye | |
| 10 | Troglodytes troglodytes | SD | April | I. ricinus | HF | Bill | |
| 11 | Turdus merula | SD | May | I. ricinus | HF | Bill | |
| Autumn | 12 | Sylvia communis | LD | August | I. ricinus | LF | Bill |
| 13† | Luscinia luscinia | LD | August | I. ricinus | FF | Ear | |
| 13† | Luscinia luscinia | LD | August | I. ricinus | HF | Bill | |
| 14 | Phoenicurus phoenicurus | LD | August | I. ricinus | HF | Bill | |
| 15 | Erithacus rubecula | SD | September | I. ricinus | U | Bill | |
| 16 | Troglodytes troglodytes | SD | September | I. ricinus | HF | Bill | |
| 17 | Turdus merula | SD | October | I. ricinus | FF | Eye | |
| 18 | Turdus merula | SD | October | I. ricinus | HF | Bill | |
| 19 | Turdus merula | SD | October | I. ricinus | HF | Eye | |
| 20 | Turdus iliacus | SD | October | N/A | HF | Bill | |
| 21 | Erithacus rubecula | SD | October | I. ricinus | FF | Bill | |
| 22 | Carduelis flammea | SD | November | N/A | U (-LF) | Bill |
§ Tick species was molecularly identified by COI and 16S.
* SD = short-distance migrant, HF = half fed, LF = little fed, FF = fully fed, U = unfed (no blood), LD = long-distance migrant (trans-Mediterranean or trans-Saharan)
† = bird that carried two Candidatus Neoehrlichia mikurensis-positive ticks.
This study contains the largest amount of ticks collected from birds that has been screened for CNM to date. The highest CNM-prevalences were recorded in nymphs collected from common redpoll (Carduelis flammea, 3/7 = 42.9%), thrush nightingale (Luscinia luscinia, 2/29 = 6.9%) and dunnock (Prunella modularis, 1/17 = 5.9%) (Table 2). The finding that CNM was recorded only from nymphs, and not from larvae, corresponds well with earlier studies and may suggest that CNM is not transovarially transmitted [8, 14, 15]. The majority of the larvae was blood-fed; they would thus presumably have acquired their infection from the bird they were feeding on. All CNM-positive ticks were identified as I. ricinus, a species which has a very wide host-feeding range. Strictly ornithophagous tick species were also found, but in low numbers and none was found positive for CNM. In this study, we found two individual birds which were infested by two CNM-positive nymphs respectively (Table 4). The first bird, a common redpoll, was infested with a total of five nymphs. Four of them were molecularly identified as I. ricinus and no sequence was obtained from the fifth, but it was morphologically identified as I. ricinus. Both CNM-positive nymphs were located at the bill of the bird, one was little fed and one was half fed (Table 4). The second bird was a thrush nightingale. It was infested with a total of four molecularly identified I. ricinus nymphs and three larvae morphologically identified as Ixodes spp. Two of the nymphs were CNM-positive; one of them fully engorged and located at the ear, the other one half fed and located at the bill of the bird. In the redpoll, the CNM-positive nymphs were attached adjacent to each other, which may indicate that CNM might be transferred among ticks co-feeding close together on the same host. The CNM-positive ticks on these two birds were all nymphs and the possibility that they independently acquired the organism as larvae cannot be ruled out. However, there are only few records of a CNM-positive larva so far [29]. Alternatively, these ticks had been infected while feeding on a systemically infected bird. However, the presence of several other CNM-negative, fully or partially engorged nymphs on the redpoll makes this assumption less likely. Unfortunately, no blood or tissue samples were collected from the tick-infested birds.
Information on possible vectors of CNM is of importance for understanding the geographical distribution of this pathogen. It is still unclear whether birds can act as reservoirs for CNM. A recent study indicated that birds bacteraemic with Rickettsia helvetica or Anaplasma phagocytophilum seem capable to infect ticks feeding on them [30].
The CNM-positive ticks were collected from 10 different bird species, all of which were predominantly ground foraging species, and thus more intensively exposed to ticks than birds within a more arboreal ecological guild. CNM was found in I. ricinus both during the avian hosts’ spring and autumn migrations. CNM-positive ticks were predominantly detected on short-distance migrant birds that spend the winter in Europe. Only few CNM-positive ticks were found on trans-Mediterranean or trans-Saharan migrants (Table 4). However, the CNM-infected ticks attached to these long-distance migrants were only collected during autumn, implying that these birds had become tick-infested in Northern Europe.
In conclusion, even though our material contained a large amount of blood-fed larvae, CNM was found only in nymphs and the prevalence in molecularly determined I. ricinus nymphs was 4.2%. This corresponds well with recent findings from Switzerland, where 3.3% of ticks collected from birds were CNM-positive, all of them nymphs [14]. This study shows that birds most likely can disperse CNM-infected ticks over large geographical areas. Different bird species may differ in their potential to disperse CNM. Their CNM dispersal potential should be influenced by their genetically determined migration patterns and other intrinsic behaviour patterns such as if they tend to feed on the ground or not.
Acknowledgments
We would like to thank the staff at Ottenby Bird Observatory for collecting samples, Matilda Karlsson and Petra Griekspoor-Berglund for skilful technical assistance and Lars Råberg at Lund University for providing a CNM-positive control extracted from rodent blood. This is contribution no. 285 from Ottenby Bird Observatory.
Data Availability
All relevant data are within the paper.
Funding Statement
Funding for this study came from the Medical Research Council of Southeast Sweden (grant numbers FORSS-307591 and FORSS-387231), Carl Trygger’s Foundation for Scientific Research, Längmanska kulturfonden, Magnus Bergvall’s Foundation for Scientific Research, Helge Ax:son Johnson's Foundation and the EU Interreg IVA project ScandTick. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kawahara M, Rikihisa Y, Isogai E, Takahashi M, Misumi H, Suto C, et al. Ultrastructure and phylogenetic analysis of 'Candidatus Neoehrlichia mikurensis' in the family Anaplasmataceae, isolated from wild rats and found in Ixodes ovatus ticks. Int J Syst Evol Microbiol. 2004;54(Pt 5):1837–43. [DOI] [PubMed] [Google Scholar]
- 2. Schouls LM, Van De Pol I, Rijpkema SG, Schot CS. Detection and identification of Ehrlichia, Borrelia burgdorferi sensu lato, and Bartonella species in Dutch Ixodes ricinus ticks. J Clin Microbiol. 1999;37(7):2215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Welinder-Olsson C, Kjellin E, Vaht K, Jacobsson S, Wenneras C. First case of human "Candidatus Neoehrlichia mikurensis" infection in a febrile patient with chronic lymphocytic leukemia. J Clin Microbiol. 2010;48(5):1956–9. 10.1128/JCM.02423-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grankvist A, Andersson PO, Mattsson M, Sender M, Vaht K, Hoper L, et al. Infections with the tick-borne bacterium "Candidatus Neoehrlichia mikurensis" mimic noninfectious conditions in patients with B cell malignancies or autoimmune diseases. Clin Infect Dis. 2014;58(12):1716–22. 10.1093/cid/ciu189 [DOI] [PubMed] [Google Scholar]
- 5. Welc-Falęciak R, Siński E, Kowalec M, Zajkowska J, Pancewicz SA. Asymptomatic "Candidatus Neoehrlichia mikurensis" Infections in Immunocompetent Humans. J Clin Microbiol. 2014;52(8):3072–4. 10.1128/JCM.00741-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Andréasson K, Jönsson G, Lindell P, Gülfe A, Ingvarsson R, Lindqvist E, et al. Recurrent fever caused by Candidatus Neoehrlichia mikurensis in a rheumatoid arthritis patient treated with rituximab. Rheumatology (Oxford). 2014. [DOI] [PubMed] [Google Scholar]
- 7. Li H, Jiang JF, Liu W, Zheng YC, Huo QB, Tang K, et al. Human Infection with Candidatus Neoehrlichia mikurensis, China. Emerg Infect Dis. 2012;18(10):1636–9. 10.3201/eid1810.120594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jahfari S, Fonville M, Hengeveld P, Reusken C, Scholte EJ, Takken W, et al. Prevalence of Neoehrlichia mikurensis in ticks and rodents from North-west Europe. Parasit Vectors. 2012;5(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maurer FP, Keller PM, Beuret C, Joha C, Achermann Y, Gubler J, et al. Close geographic association of human neoehrlichiosis and tick populations carrying "Candidatus Neoehrlichia mikurensis" in eastern Switzerland. J Clin Microbiol. 2013;51(1):169–76. 10.1128/JCM.01955-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Richter D, Kohn C, Matuschka FR. Absence of Borrelia spp., Candidatus Neoehrlichia mikurensis, and Anaplasma phagocytophilum in questing adult Dermacentor reticulatus ticks . Parasitology research. 2013;112(1):107–11. 10.1007/s00436-012-3110-8 [DOI] [PubMed] [Google Scholar]
- 11. Vayssier-Taussat M, Le Rhun D, Buffet JP, Maaoui N, Galan M, Guivier E, et al. Candidatus Neoehrlichia mikurensis in bank voles, France. Emerg Infect Dis. 2012;18(12):2063–5. 10.3201/eid1812.120846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Andersson M, Råberg L. Wild Rodents and Novel Human Pathogen Candidatus Neoehrlichia mikurensis, Southern Sweden. Emerg Infect Dis. 2011;17(9):1716–8. 10.3201/eid1709.101058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hasle G. Transport of ixodid ticks and tick-borne pathogens by migratory birds. Front Cell Infect Microbiol. 2013;3:48 10.3389/fcimb.2013.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lommano E, Dvorak C, Vallotton L, Jenni L, Gern L. Tick-borne pathogens in ticks collected from breeding and migratory birds in Switzerland. Ticks Tick Borne Dis. 2014;5(6):871–82. 10.1016/j.ttbdis.2014.07.001 [DOI] [PubMed] [Google Scholar]
- 15. Spitalská E, Literák I, Sparagano OA, Golovchenko M, Kocianová E. Ticks (Ixodidae) from passerine birds in the Carpathian region. Wien Klin Wochenschr. 2006;118(23–24):759–64. [DOI] [PubMed] [Google Scholar]
- 16. Movila A, Alekseev AN, Dubinina HV, Toderas I. Detection of tick-borne pathogens in ticks from migratory birds in the Baltic region of Russia. Med Vet Entomol. 2012. [DOI] [PubMed] [Google Scholar]
- 17. Arthur DR. British ticks. London: Butterworths; 1963. [Google Scholar]
- 18. Filippova NA. Ixodid ticks of the subfamily Ixodinae 4th Edition ed. Leningrad: Nauka; 1977. 396 p. [Google Scholar]
- 19. Hillyard PD. Ticks of North-West Europe: Keys and notes for identification of the species Synopses of the British Fauna. New Series no 52 Edited by Banks R.S.K. and Crothers J. H.. Shrewsbury: The Linnean Society of London and The Estuarine and Coastal Sciences Association; 1996. [Google Scholar]
- 20. Heylen D, De Coninck E, Jansen F, Madder M. Differential diagnosis of three common Ixodes spp. ticks infesting songbirds of Western Europe: Ixodes arboricola, I. frontalis and I. ricinus . Ticks Tick Borne Dis. 2014;5(6):693–700. 10.1016/j.ttbdis.2014.05.006 [DOI] [PubMed] [Google Scholar]
- 21. Lv J, Wu S, Zhang Y, Chen Y, Feng C, Yuan X, et al. Assessment of four DNA fragments (COI, 16S rDNA, ITS2, 12S rDNA) for species identification of the Ixodida (Acari: Ixodida). Parasit Vectors. 2014;7:93 10.1186/1756-3305-7-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Andersson M, Bartkova S, Lindestad O, Raberg L. Co-infection with 'Candidatus Neoehrlichia mikurensis' and Borrelia afzelii in Ixodes ricinus ticks in southern Sweden. Vector borne and zoonotic diseases. 2013;13(7):438–42. 10.1089/vbz.2012.1118 [DOI] [PubMed] [Google Scholar]
- 23. Wilhelmsson P, Lindblom P, Fryland L, Ernerudh J, Forsberg P, Lindgren PE. Prevalence, Diversity, and Load of Borrelia species in Ticks That Have Fed on Humans in Regions of Sweden and Åland Islands, Finland with Different Lyme Borreliosis Incidences. PLoS One. 2013;8(11):e81433 10.1371/journal.pone.0081433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lv J, Wu S, Zhang Y, Zhang T, Feng C, Jia G, et al. Development of a DNA barcoding system for the Ixodida (Acari: Ixodida). Mitochondrial DNA. 2014;25(2):142–9. 10.3109/19401736.2013.792052 [DOI] [PubMed] [Google Scholar]
- 25. Zhang RL, Zhang B. Prospects of using DNA barcoding for species identification and evaluation of the accuracy of sequence databases for ticks (Acari: Ixodida). Ticks Tick Borne Dis. 2014;5(3):352–8. 10.1016/j.ttbdis.2014.01.001 [DOI] [PubMed] [Google Scholar]
- 26. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–25. [DOI] [PubMed] [Google Scholar]
- 27. Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27(2):221–4. 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- 28. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Derdáková M, Václav R, Pangrácova-Blaňárová L, Selyemová D, Koči J, Walder G, et al. Candidatus Neoehrlichia mikurensis and its co-circulation with Anaplasma phagocytophilum in Ixodes ricinus ticks across ecologically different habitats of Central Europe. Parasit Vectors. 2014;7:160 10.1186/1756-3305-7-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hornok S, Kováts D., Csörgo T., Meli M.L., Gönczi E., Hadnagy Z., Takács N., Farkas R., Hofmann-Lehmann R. Birds as potential reservoirs of tick-borne pathogens: first evidence of bacteraemia with Rickettsia helvetica . Parasit Vectors. 2014;7(128). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.