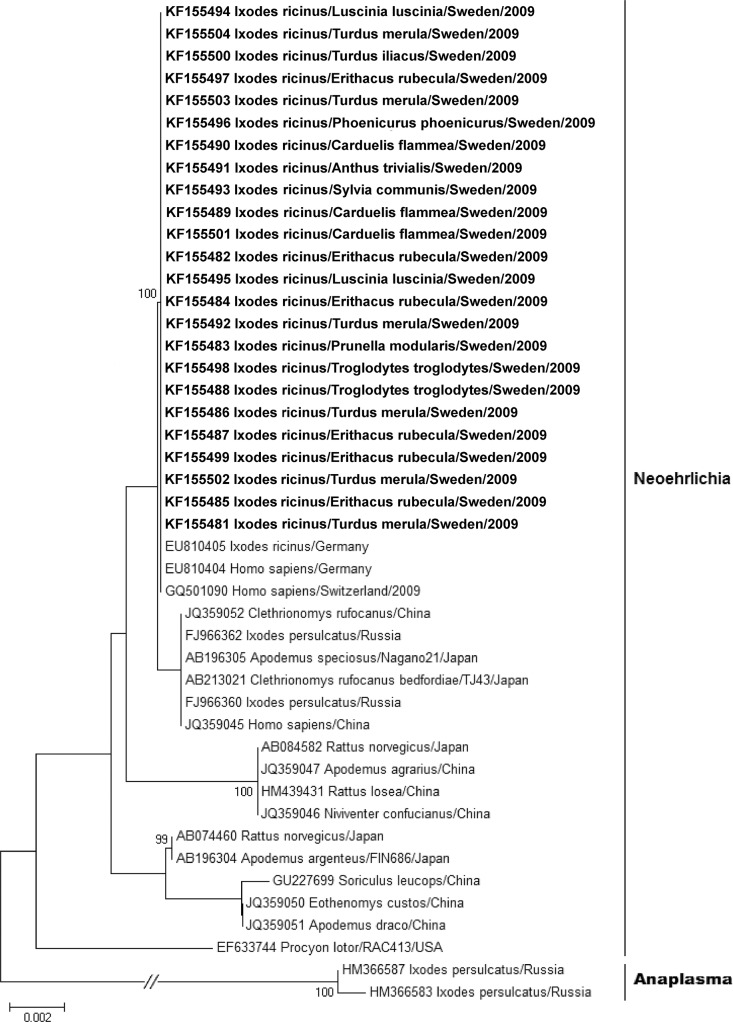
Fig 1. Phylogenetic relatedness of Candidatus Neoehrlichia mikurensis sequences.
The tree is based on the partial nucleotide sequence of the 16S rRNA gene of CNM and Anaplasma phagocytophilum. Bacterial sequences generated in this study (boldface) and previously reported sequences are denoted with GenBank accession number followed by information regarding sequence source. The tree was inferred using the neighbour joining method implemented in the SeaView software version 4 [27], utilizing 1000 bootstrap replications to determine support for inferred nodes. The tree was visualized using the MEGA software version 5.2 [28], and statistical supports of > 95% for inferred nodes are displayed. In the mid-point rooted tree, the branch between the CNM and the Anaplasma clade has been truncated in order to limit the tree size. The scale bar represents the number of substitutions per site. The CNM sequences were all 100% identical to each other as well as to three other European strains, including those detected in humans, and therefore appear in a single clade.