Fig. 1.
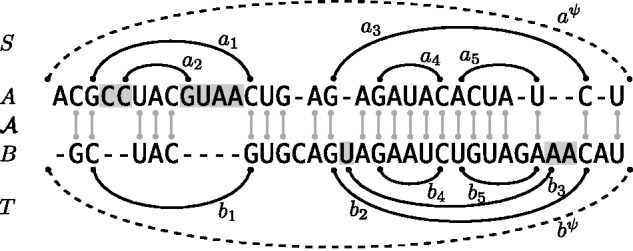
Example alignment of two RNA sequences A and B together with (non-crossing) structures and . We highlight the positions in the loop closed by a1 and in the loop closed by b2. The base pair a1 is the parent of the highlighted positions in A and of a2. The base pair a1 closes a 2-loop; a2, a 1-loop and a3, a multiloop. The latter is a 3-loop, since a3 has two inner base pairs (a4 and a5.) Note that the structure alignment triple covers the external base pairs and as well as the inner base pairs of the two multiloops. Finally, deletes the entire 2-loop of a1 and inserts the entire 2-loop of b2
