Figure 1.
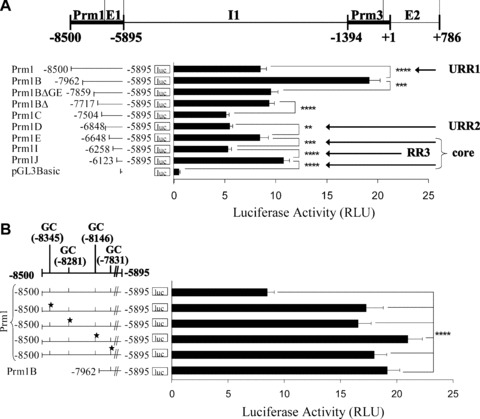
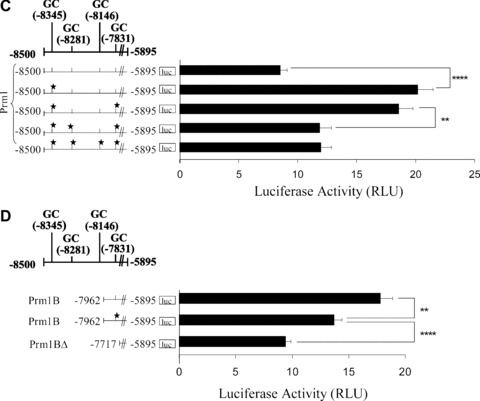
Effect of 5′ deletions on Prm1-directed gene expression and identification of GC elements within the –8500 to –7962 region of Prm1. (A): Schematic of the human TP gene spanning nucleotides –8500 to +786 encoding Prm1 (–8500 to –5895), Prm3, exon (E)1, intron (I)1 and E2, where nucleotide +1 represents the translational start site (ATG). pGL3Basic plasmids (2 μg) encoding Prm1 (–8500 to –5895) or its 5′ deletion fragments Prm1B (–7962), Prm1BΔGata/Ets (–7859), Prm1BΔ (–7717), Prm1C (–7504), Prm1D (–6848), Prm1E (–6648), Prm1I (–6258), Prm1J (–6123) or, as a control, pGL3Basic were co-transfected with pRL-TK into HEL 92.1.7 cells. Mean firefly relative to renilla luciferase activity was expressed in arbitrary relative luciferase units (RLU ± S.E.M.; n= 6). (B), (C) and (D): GC elements containing putative overlapping WT1/Egr1/Sp1 binding sites within Prm1, where the 5′ nucleotide is indicated and the star symbol signifies mutated elements. pGL3Basic plasmids (2 μg) encoding: (B) Prm1, Prm1GC*(–8345), Prm1GC*(–8281), Prm1GC*(–8146), Prm1GC*(–7831) and Prm1B; (C) Prm1, Prm1GC*(–8345), Prm1GC*(–8345,–7831), Prm1GC*(–8345,–8281,–7831) and Prm1GC*(–8345,–8281,–8146,–7831); or (D) Prm1B, Prm1BGC*(–7831) and Prm1BΔ were co-transfected with pRL-TK into HEL cells. Luciferase activity was expressed as mean firefly relative to renilla luciferase activity (RLU ± S.E.M.; n= 8).
