DAZL-positive cells are highly hydroxymethylated and reprogram efficiently to a naïve pluripotent state
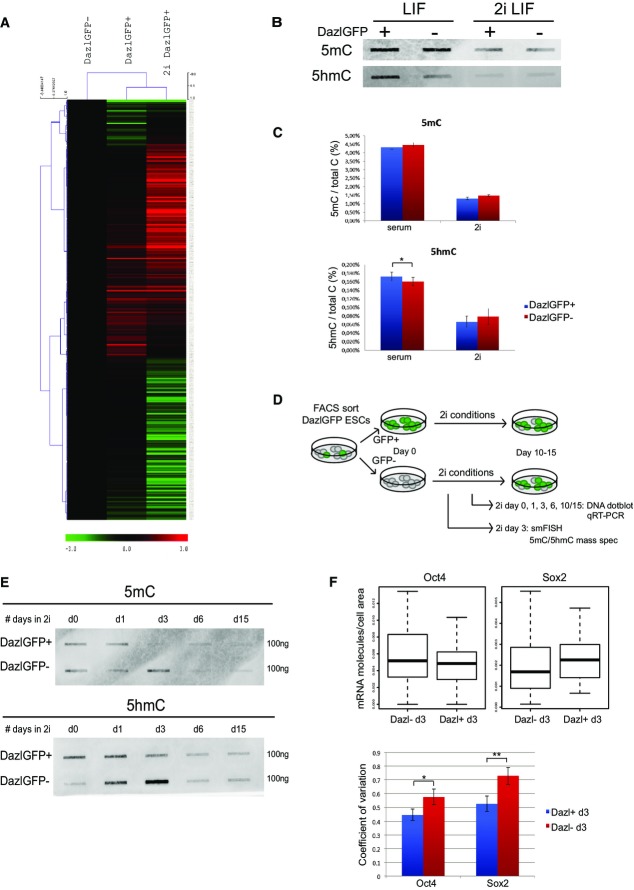
Proteomic analysis of genes expressed in Dazl-negative ESCs, Dazl-positive ESCs, and Dazl-positive ESCs cultured for 12 days in 2i conditions.
Dot-blot analysis for 5meC and 5hmeC in Dazl-positive and Dazl-negative ESCs FACS-sorted from serum-cultured cells or sorted from ESCs cultured in 2i + LIF for 12 days.
Mass spectrometric measurement of global 5mC and 5hmC levels in DazlGFP+ and DazlGFP− ESCs in serum as well as in 2i conditions. P-values were calculated using paired t-test. *P < 0.05. Error bars indicate s.e.m. values of ≥ 3 biological replicates.
Schematic overview of experimental setup: DazlGFP+ ESCs and DazlGFP− ESCs are FACS-sorted from serum-cultured ESCs followed by culture of each population in 2i conditions. Different time points during the transition to a naïve pluripotent state were analyzed for gene expression and DNA methylation.
Dot-blot analysis for 5meC and 5hmeC in Dazl-positive and Dazl-negative cells FACS-sorted at day 0 followed by culture in 2i + LIF conditions for several days.
The upper panel shows a boxplot with the distribution of single Oct4 and Sox2 mRNA molecules per cell area in > 50 individual ESCs that were sorted for Dazl-GFP+ and Dazl-GFP− followed by 3 days of culture in 2i conditions. The lower panel shows the coefficient of variation (or normalized variance) of the single Oct4 and Sox2 transcripts within each cell population. *P < 0.05, **P < 0.001. Error bars indicate s.d. values of 100,000 times in silico random sampling of 50 cells per condition.