Figure 1.
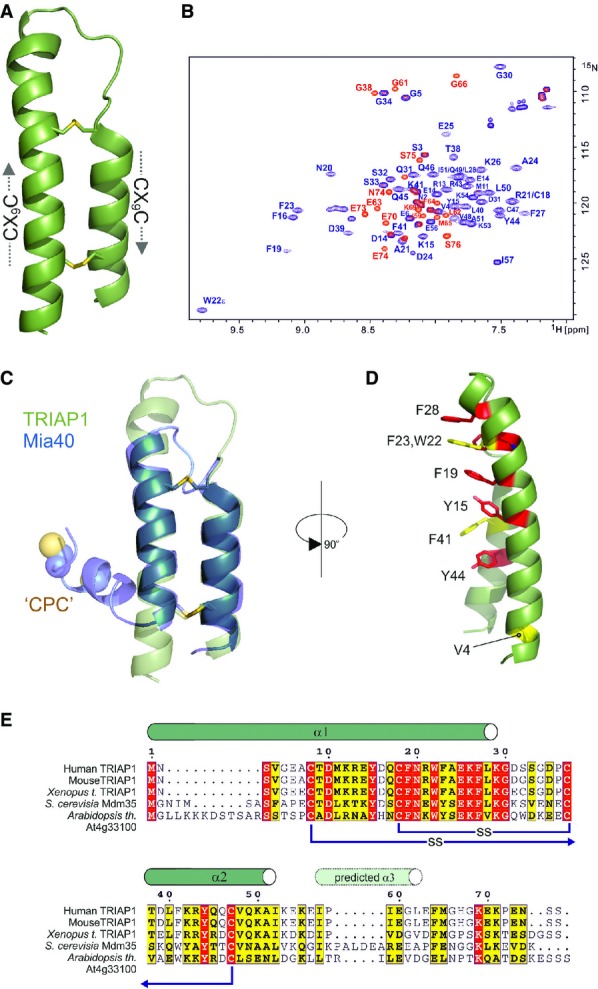
- Cartoon representation in green for the crystal structure of TRIAP1 showing the location of the disulphide bonds (yellow) and the twin CX9C motifs.
- 1H-15N HSQC NMR spectrum of TRIAP1ΔC (blue) overlaid on a single 1H-15N HSQC NMR spectrum of full-length TRIAP1 from a 15N-transverse relaxation measurement series (red). The relaxation delay for the time point in the 15N-transverse relaxation experiment was set such that all 15N-signals for the folded coiled coil have decayed and therefore would not be observed in the spectrum. Remaining signals observed represent amides with slow transverse relaxation and therefore are highly dynamic and disordered. Assignment of the 1H-15N HSQC spectra reveals that this flexible region is localised to the C-terminus of full-length TRIAP1.
- Cartoon representation for the superposition of TRIAP1 with the solution structure of Mia40 revealing topological similarity of the twin CX9C-coiled coil domain.
- Conserved hydrophobic stripe on the surface of TRIAP1 comprising conserved aromatic residues. The conserved V4 residue in the N-terminus of helix I is also shown. Backbone of TRIAP1 is shown as a cartoon and stacked aromatic side chains labelled in stick representation. Shading of side chains according to alignment shown in (E).
- Protein sequence alignment for TRIAP1, Mdm35 and selected homologues. Positions of experimentally determined and predicted α-helices are indicated above the alignment. Disulphide bond connectivities are also indicated. Red shading indicates invariant residues across the homologues, and yellow shading indicates locations where there are conserved residues in three homologues.
