Figure 4.
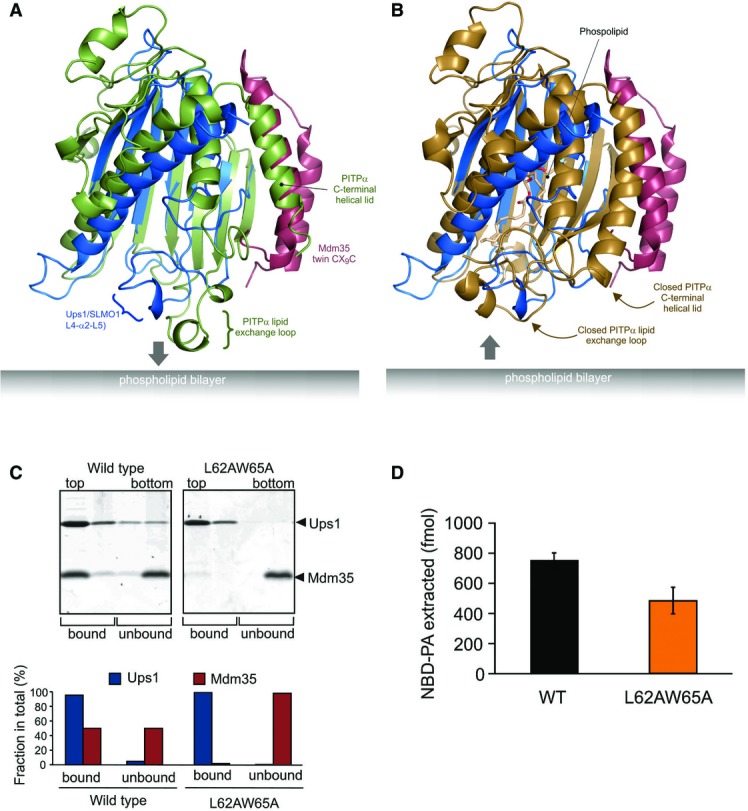
- Cartoon representation for the superposition of the modelled Mdm35-Ups1 structure (blue and red, respectively) with apo mouse PITPα (pdb: 1KCM; green). The identified lipid exchange loop in PITPα and equivalent region in Ups1 (L4-a2-L5) are indicated.
- Cartoon representation for the superposition of the modelled Mdm35-Ups1 structure (blue and red, respectively) with phosphatidylcholine-bound rat PITPα (pdb: 1T2Z; brown). Conformational changes in lipid exchange loop and the C-terminal helical lid (indicated by arrows) close the structure and cap the bound phospholipid.
- Liposome binding. Purified Mdm35-Ups1 complex or its mutant variant was incubated with liposomes composed of DOPC/POPE/DOPA (50/30/20%), and binding was assessed by flotation of liposomes in a sucrose gradient. Upper panel: fractions after sucrose gradient were analysed by SDS–PAGE and CBB staining. All liposomes were recovered in the upper two fractions. Lower panel: quantification of Ups1 and Mdm35. Signals in upper two fractions (bound) or lower two fractions (unbound) were quantified and represented as a fraction of total signals of four fractions.
- NBD-PA extraction. Purified Mdm35-Ups1 complexes (80 nM) were incubated with liposomes (4 μM) composed of DOPC/POPE/NBD-PA/Rhod-PE (50/43/5/2%) filled with 12.5% sucrose. After incubation at 25°C for 2 min, liposomes were sedimented by an ultracentrifugation step (200,000× g, 30 min) and NDB fluorescence in the supernatant fraction was quantified using standard probes of NBD-PA. Columns and error bars indicate the mean ± SD. n = 3.
