Figure 5.
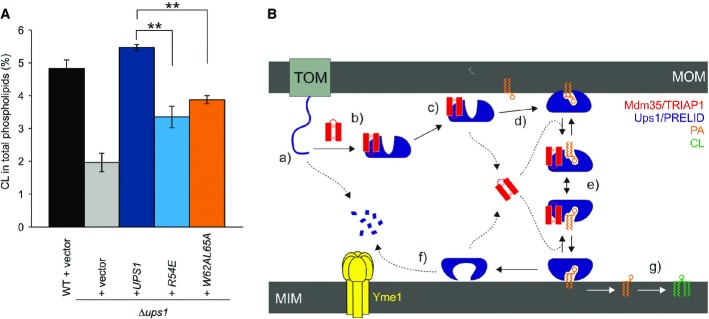
- Restoration of CL levels by plasmid-encoded Ups1 and its mutant variants. Δups1 cells carrying empty vector (YCplac111ADH, vector) or a plasmid encoding indicated Ups1 variants were grown to logarithmic phase in YP medium supplemented with 2% galactose. Cells were collected and subjected to lipid extraction and phospholipidome analysis. CL levels were represented as a proportion in total phospholipids. Wild-type cells carrying empty vector were analysed as a control. Columns and error bars indicate the mean ± SD. n = 3. Student's t-test was used to calculate P-values. **P < 0.01.
- Proposed mechanism of phosphatidic acid (PA) transport by PRELI-like domains. Phospholipid transport between mitochondrial inner and outer membranes (MIMs & MOMs) catalysed by TRIAP1/Mdm35-PRELID complexes: (a) import of PRELID and degradation by mitochondrial proteases if no complex formed; (b) folding of PRELI-like domain on the TRIAP1/Mdm35 twin coiled-coiled CX9C motif; (c) donor bilayer (MOM) binding and preloading by the complex; (d) PA loading and dissociation of TRIAP1/Mdm35; (e) recapture of loaded PRELID by TRIAP1/Mdm35, donor bilayer binding and delivery of PA; (f) degradation of PRELID at the MIM; and (g) biosynthesis of cardiolipin (CL) from PA.
